Brian Snyder and Christopher H. Contag, from Michigan State University, discuss engineering interkingdom communication, which is not for palace intrigue, but for next-generation therapeutic approaches they argue
Redefining symbiosis
Our gut microbiota, once seen as ‘harmless passengers’, is now redefined as a vital symbiotic organ integral to sustaining human life. This microbiome consists of an estimated 1×1014 microbial symbionts. With an approximate 1:1 ratio of human to bacterial cells, it is the largest “organ” in the human body by cell count.
Facilitating digestion, nutrient absorption, immune responses, and metabolic function, microbial symbionts are crucial for maintaining human health. Even minor changes in the composition of our microbiome have been implicated in a myriad of diseases, including cancers, metabolic diseases, autoimmune disorders, and neurological conditions (Clemente, Ursell, et al. 2012; Gomaa 2020).
Comparable to mammalian organs, the biological functions of our microbiota do not occur in isolation from the rest of the body; there is a continuous and bidirectional communication occurring between human cells and bacterial cells. This dynamic exchange of molecular emissaries ensures our human body and microbiota work collaboratively, maintaining a balance, i.e., homeostasis. This phenomenon is colloquially referred to as interkingdom communication as humans and bacterial cells are classified under distinct and separate kingdoms of life, Animalia and Eubacteria – cellular life across all six kingdoms of life interacts and communicates bidirectionally. (Celluzzi and Masotti 2016, Teng, Ren et al. 2018).
Exploiting interkingdom communication
These interkingdom communications have emerged at the forefront of research in life sciences. By determining the specific molecular elements that act as molecular emissaries between kingdoms, we can exploit these mechanisms to develop new therapeutic strategies and discover novel therapeutic molecules. Molecules, including lipids, proteins, and nucleic acids, have been shown to facilitate cross-talk between bacteria and mammalian cells (Enaud, Prevel, et al. 2020, Diaz-Garrido, Badia et al. 2021), and disruption of these molecular channels of communication often leads to disease. (Enaud, Prevel, et al. 2020; Ormseth, Wu et al. 2020; Sarshar, Scribano, et al. 2020).
RNA has frequently been found taking on an emissarial role in interkingdom communication (Celluzzi and Masotti 2016; Ormseth, Wu et al. 2020; Sarshar, Scribano, et al. 2020; Stanton 2021). Once constrained to the designation of acting only as a message between DNA and protein, our current understanding of RNA goes well beyond this dogma. We now know that noncoding RNAs have many physiologic roles, including regulation, defense and communication (Gottesman 2005, Cech and Steitz 2014). Notably, since the first FDA- approved RNA therapeutic was released in 2018, there have been four additional FDA-approved RNA-based therapeutics on the market.
RNA is arguably the perfect molecular conduit for communicating between different kingdoms of life owing to a few unique characteristics. RNA is a universal language among all life, meaning every living cell is equipped to interact with and respond to it as a signaling molecule.
RNA is also inherently information-rich; its sequence, base modifications, and structure is diverse while also being energetically “cheaper” and quicker to synthesize in comparison to proteins. Its inherent instability ensures that communiques are “written in disappearing ink,” which ensures that messages don’t outlast the intended signal.
Lastly, the evolution barrier is an order of magnitude less than that of protein; incurring a mutation within the DNA inevitably alters the RNA sequence but does not guarantee an alteration to the protein; thus, the threshold to evolutionarily adapt through RNA is lower than via its protein counterpart.
Endosymbionts and engineered interkingdom communiques
Unlike the majority of symbionts which are extracellular, a subset of symbionts resides intracellularly, or within the confines of another cell, called endosymbionts. Mitochondria, now bonified organelles, originated when primitive eukaryotic cells engulfed a proteobacteria approximately 2 billion years ago – a naturally occurring endosymbiosis (Martin, Garg et al. 2015). Over time, evolution selected for an interdependent relationship resulting in a reduction of the mitochondrial genome as it evolved from an endosymbiont into an essential organelle.
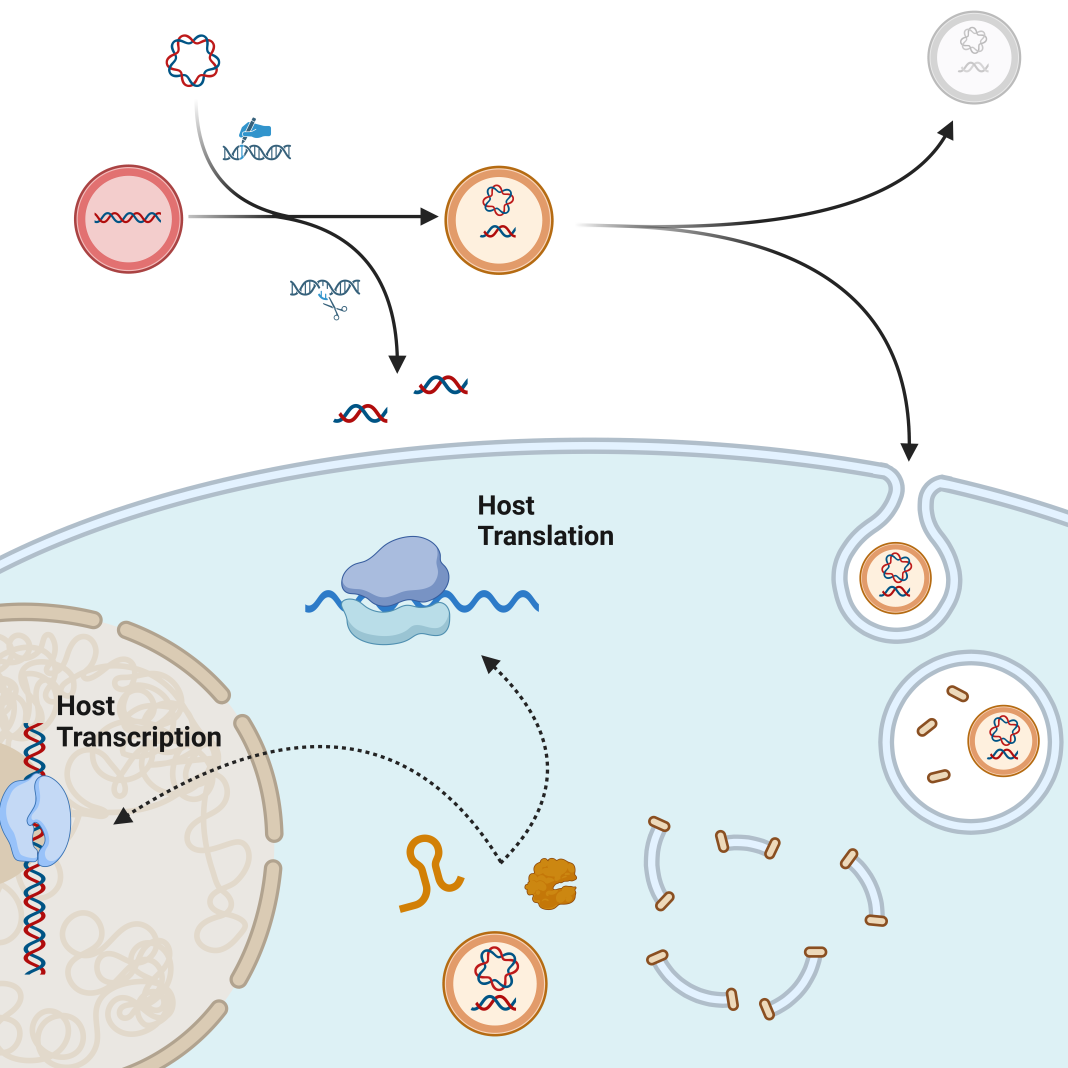
Inspired by the evolutionary origins of mitochondria, our group and others have set out to mimic mitochondria by creating engineered endosymbionts (EES) as next-generation cellular control modules and therapeutics. Naturally occurring endosymbionts are teaching us how to engineer innocuous bacteria to persist within target cells and produce cellular communicators when signaled to do so and elicit a desired outcome.
Bacterial endosymbionts have a tremendous capacity to encode and deliver molecular signals with a multitude of regulatory pathways to provide exquisite control over complex signaling networks. Complex biological processes that lead to cancer or autoimmunity, or that control tissue regeneration after organs are damaged or lost, could be controlled by molecular communiques delivered from EES to cells of another kingdom. As such, EES presents a paradigm shift in how we can control normal and disease processes.
Creating an innocuous, inert, and sustained EES
Concurrent with interkingdom communication, interkingdom warfare naturally developed when biological kingdoms diverged. Bacteria have constantly evolved mechanisms to invade and hijack mammalian cells, and mammalian systems have evolved multiple mechanisms of sensing and eradicating the invaders. The first challenge to creating EES is to learn from successful endosymbionts to engineer harmonious relationships that do not set off alarms to trigger immune responses.
Taking another lesson from mitochondria, a minimal genome is a key characteristic when creating an EES, and genes have been eliminated from bacterial EES to minimize the genome (Mehta, Ko et al. 2019; Madsen, Makela, et al. 2022). Generating an effective EES will require engineering a dependency of the EES on the recipient cell for an engineered symbiosis.
Signaling induced drug production
The next challenge is to have the EES produce molecular signals that are delivered to mammalian cells in response to external signals. It has already been demonstrated that engineered bacterium can be directed to produce and secrete therapeutic proteins (Jin, Liu et al. 2018, Bracha, Johnson et al. 2024). Transcription factors are proteins used by all cells to control gene expression; however, each species tends to speak its own unique molecular language.
Bacteria can, however, be engineered to make mammalian transcription factors. In one example, a strain of P. aeruginosa was engineered to inject these factors, using a “molecular needle”, into stem cells to inform them to differentiate into heart cells (Jin, Liu et al. 2018). This interkingdom communication was not from an endosymbiont but rather from an extracellular bacteria.
Similarly, we have demonstrated that we can engineer EES to produce transcription factors from inside mammalian cells and drive cellular programs that stoke a proinflammatory signaling cascade to effectively kill cancer cells (Madsen, Makela, et al. 2022). In addition to these protein- based interkingdom communiques, an RNA-based signal has also been produced by an internalized bacterium (Zhao, Zhou et al. 2017). Intracellular localization helps the EES evade the immune system to deliver molecular mediators that change cellular fates and function.
In addition to having an enormous capacity for carrying molecular cargo, EES, as a living therapeutic, it can be controlled and directed from outside the body. This can be accomplished by introducing a signaling molecule such as a unique sugar (Madsen, Makela, et al. 2022) or using an external source of energy such as a magnetic field or focused ultrasound (Greeson, Madsen, et al. 2022).
Hypothetically a genetic timer could also be introduced such that once the EES is successfully internalized within a desired cell, a cell-specific signal can trigger the EES to produce the therapeutics and remain as an intracellular endosymbiont. This new field of engineered endosymbionts is in its infancy; however, it offers a dramatic paradigm shift in how to maintain human health. The potential of this therapeutic platform is massive and indisputable, enabling the development of new biological therapeutics and can be multiplexed, regulated, and directed to guide mammalian biology toward health.
Reference
- Bracha, S., H. J. Johnson, N. A. Pranckevicius, F. Catto, A. E. Economides, S. Litvinov, K. Hassi, M. T. Rigoli, C. Cheroni, M. Bonfanti, A. Valenti, S. Stucchi, S. Attreya, P. D. Ross, D. Walsh, N. Malachi, H. Livne, R. Eshel, V. Krupalnik, D. Levin, S. Cobb, P. Koumoutsakos, N. Caporale, G. Testa, A. Aguzzi, A. A. Koshy, L. Sheiner and O. Rechavi (2024). “Engineering Toxoplasma gondii secretion systems for intracellular delivery of multiple large therapeutic proteins to neurons.” Nat Microbiol 9 (8): 2051-2072.
- Cech, T. R. and J. A. Steitz (2014). “The noncoding RNA revolution-trashing old rules to forge new ones.” Cell 157 (1): 77-94.
- Celluzzi, A. and A. Masotti (2016). “How Our Other Genome Controls Our Epi-Genome.” Trends Microbiol 24 (10): 777-787.
- Clemente, J. C., L. K. Ursell, L. W. Parfrey and R. Knight (2012). “The impact of the gut microbiota on human health: an integrative view.” Cell 148 (6): 1258-1270.
- Diaz-Garrido, N., J. Badia and L. Baldoma (2021). “Microbiota-derived extracellular vesicles in interkingdom communication in the gut.” J Extracell Vesicles 10 (13): e12161.
- Enaud, R., R. Prevel, E. Ciarlo, F. Beaufils, G. Wieers, B. Guery and L. Delhaes (2020). “The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks.” Front Cell Infect Microbiol 10: 9.
- Gomaa, E. Z. (2020). “Human gut microbiota/microbiome in health and diseases: a review.” Antonie Van Leeuwenhoek 113 (12): 2019-2040.
- Gottesman, S. (2005). “Micros for microbes: non-coding regulatory RNAs in bacteria.” Trends Genet 21 (7): 399-404.
- Greeson, E. M., C. S. Madsen, A. V. Makela and C. H. Contag (2022). “Magnetothermal Control of Temperature-Sensitive Repressors in Superparamagnetic Iron Nanoparticle-Coated Bacillus subtilis.” ACS Nano 16 (10): 16699-16712.
- Jin, Y., Y. Liu, Z. Li, K. Santostefano, J. Shi, X. Zhang, D. Wu, Z. Cheng, W. Wu, N. Terada, S. Jin and F. Bai (2018). “Enhanced differentiation of human pluripotent stem cells into cardiomyocytes by bacteria-mediated transcription factors delivery.” PLoS One 13 (3): e0194895.
- Madsen, C. S., A. V. Makela, E. M. Greeson, J. W. Hardy and C. H. Contag (2022). “Engineered endosymbionts that alter mammalian cell surface marker, cytokine and chemokine expression.” Commun Biol 5 (1): 888.
- Martin, W. F., S. Garg and V. Zimorski (2015). “Endosymbiotic theories for eukaryote origin.” Philos Trans R Soc Lond B Biol Sci 370 (1678): 20140330.
- Mehta, A. P., Y. Ko, L. Supekova, K. Pestonjamasp, J. Li and P. G. Schultz (2019). “Toward a Synthetic Yeast Endosymbiont with a Minimal Genome.” J Am Chem Soc 141 (35): 13799-13802.
- Ormseth, M. J., Q. Wu, S. Zhao, R. M. Allen, J. Solus, Q. Sheng, Y. Guo, F. Ye, M. Ramirez-Solano, S. L. Bridges, J. R. Curtis, K. Vickers and C. M. Stein (2020). “Circulating microbial small RNAs are altered in patients with rheumatoid arthritis.” Ann Rheum Dis 79( 12): 1557-1564.
- Sarshar, M., D. Scribano, C. Ambrosi, A. T. Palamara and A. Masotti (2020). “Fecal microRNAs as Innovative Biomarkers of Intestinal Diseases and Effective Players in Host-Microbiome Interactions.” Cancers (Basel) 12 (8).
- Stanton, B. A. (2021). “Extracellular Vesicles and Host-Pathogen Interactions: A Review of Inter-Kingdom Signaling by Small Noncoding RNA.” Genes (Basel) 12(7).
- Teng, Y., Y. Ren, M. Sayed, X. Hu, C. Lei, A. Kumar, E. Hutchins, J. Mu, Z. Deng, C. Luo, K. Sundaram, M. K. Sriwastva, L. Zhang, M. Hsieh, R. Reiman, B. Haribabu, J. Yan, V. R. Jala, D. M. Miller, K. Van Keuren-Jensen, M. L. Merchant, C. J. McClain, J. W. Park, N. K. Egilmez and H. G. Zhang (2018). “Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota.” Cell Host Microbe 24 (5): 637-652 e638.
- Zhao, C., Z. Zhou, T. Zhang, F. Liu, C. Y. Zhang, K. Zen and H. Gu (2017). “Salmonella small RNA fragment Sal-1 facilitates bacterial survival in infected cells via suppressing iNOS induction in a microRNA manner.” Sci Rep 7 (1): 16979.

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.