Here, David Ussery from the Department of BioMedical Informatics, UAMS, details E. coli genome diversity, big data, and messy biology. New methods, we discover, allow for the comparison of millions of bacterial genomes in a few days and the confident assignment of taxonomic clusters
Escherichia coli was first described in 1885 (1) and is one of the best-studied model organisms as both a commensal (2) and a pathogen. (3) The diversity of E. coli is enormous. We have compared more than 10,000 E. coli genomes and found more than a hundred thousand different gene families distributed in the ‘pan-genome’. (4) Since then, the number of sequenced E. coli genomes has continued to grow and is now close to a million.
Where are we now? E. coli diversity is vast!
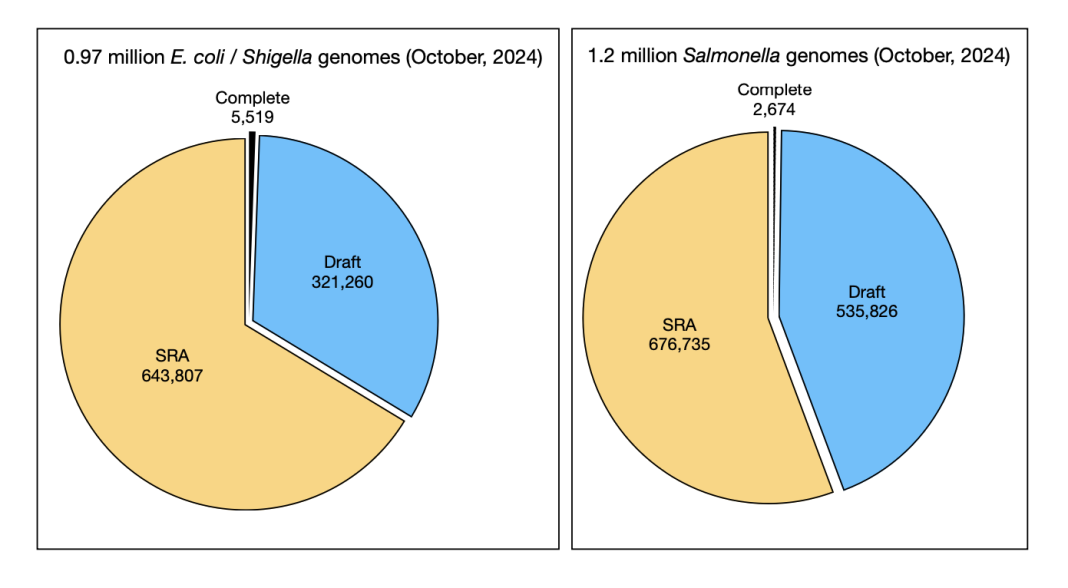
At the time of writing this article (October, 2024), there are just under a million E. coli genome sequences available in the NCBI database, with more than 5,000 complete E. coli/ Shigella genomes, and another 321,260 draft genomes; in addition, there are nearly twice as many genomes in the Sequence Read Archive (SRA) database, bringing the total to more than 970,000 E. coli genomes, as shown in Figure 1; there are likely more than a million different gene families found in this set of E. coli genomes, although any individual E. coli contains about 5,000 genes.
How did we get here? A brief history of E. coli genomics Sometimes, fact can be stranger than fiction. Who would have thought that it is possible for one bacterial species to have more than a million different proteins? The first E. coli genome was sequenced in 1997 from a laboratory strain, with about 4,300 genes. (5) At the time, the vast genomic diversity of this organism was not appreciated. Around this time, we developed the ‘genome atlas’ for E. coli, a way to view the entire chromosome as one circular plot. (6,8)
A few years later, the second E. coli genome sequence (this time from a pathogenic strain), with about 5,500 genes; many were surprised that this contained more than a thousand extra genes not found in the first E. coli genome. (9) A microarray with four different E. coli genomes soon became available (10), followed by microarrays with seven (11) and then thirty-two E. coli genomes (12).
A comparison of 61 E. coli genomes found a pan-genome with more than 15,000 different E. coli gene families, and a core of only 960 gene families (13). Two years later, the number of E. coli genomes had more than doubled (14), and as more genomes were compared, we realized that the presence of low-quality draft genomes was causing the core to drop to near zero, so we re-defined ‘core’ genes as being present in at least 95% of the genomes. This resulted in a stable core of roughly 3,000 gene families found in 400 E. coli genomes (15), and then 2,000 E. coli genomes (16).
Since then, the number of E. coli genomes (and the diversity) has steadily grown with time. More recently, we have compared more than 10,000 E. coli genomes and found a core of about 2,800 gene families, and most of the genomes are clustered into 14 phylogroups that include many Shigella species (4).
Where are we headed in the future of ‘big data in biology’?
E. coli genomics now requires dealing with large amounts of messy data. I see two areas of concern: scalable and reproducible methods for comparing millions of genomes with consistent organism names and getting the organism names right. Making predictions is difficult – especially predictions about the future. The Enterobacteriaceae family (“E. coli and friends”) has been well-studied and heavily sequenced; there are more than 1.2 million Salmonella genomes, as well as more than another hundred thousand other genomes from the Enterobacteriaceae family.
Currently, we have found that the Mash program (17) seems to give reproducible and consistent yields and scales well. For example, we have recently clustered more than a million Salmonella genomes (see Figure 1) into a dozen phylogroups, as we’ve done for 100,000 E. coli genomes (4), which took less than a week, using a small cluster. Having said that, getting the data from the SRA is not easy.
Originally, we estimated it would take about 14 months to download the 600,000 Salmonella genomes in the SRA. Fortunately, another research group had assembled all the bacterial genomes (18), and we could download all the Salmonella genomes in less than one day. However, we found that about 8% of the ‘Salmonella’ genomes clustered with E.coli (Sudip Panday and Dave Ussery, unpublished results).
Since the time of Aristotle, biologists have been naming things, and more recently, there has been a rush to assign names to bacteria based on their genome sequence. New names for bacterial species are being proposed daily, and about 80% of the names of bacterial species have been changed in the past two years . In early versions of the Genome Taxonomy Database, many stains of E. coli were given new names (19). For example, E. coli K-12 was renamed ‘Escherichia flexneri’, since this sequence was close to Shigella flexneri. The idea of changing the name of one of the most well-known and studied organisms was met with resistance, and for now the ‘rose’ of E. coli is still being called a ‘rose’ (20).
References
- Escherich T, “The intestinal bacteria of the neonate and breast-fed infant. 1885”, Rev Infect Disease, 11:352-356, (1989). https://doi.org/10.1093/clinids/11.2.352 PubMed PMID: 2649968.
- Blount ZD, “The unexhausted potential of E. coli”, Elife, 4. Epub 20150325 (2015). doi: https://doi.org/10.7554/eLife.05826. PubMed PMID: 25807083; PMCID: PMC4373459.
- Denamur E, Clermont O, Bonacorsi S, Gordon D, “The population genetics of pathogenic Escherichia coli”, Nature Reviews Microbiology, 19:37-54, (2021). Epub 20200821. https://doi.org/10.1038/s41579-020-0416-x PubMed PMID: 32826992.
- Abram K, Udaondo Z, Bleker C, Wanchai V, Wassenaar TM, Robeson MS, 2nd, Ussery DW, “Mash-based analyses of Escherichia coli genomes reveal 14 distinct phylogroups”, Commun Biol. 2021;4(1):117. Epub 20210126. https://doi.org/10.1038/s42003-020-01626-5 PubMed PMID: 33500552; PMCID: PMC7838162.
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y, “The complete genome sequence of Escherichia coli K-12”, Science, 277:1453-1462 (1997). https://doi.org/10.1126/science.277.5331.1453 PubMed PMID: 9278503.
- Jensen LJ, Friis C, Ussery DW, “Three views of microbial genomes”, Research in Microbiology, 150:773-777, (1999). https://doi.org/10.1016/s0923-2508(99)00116-3 PubMed PMID: 10673014.
- Pedersen AG, Jensen LJ, Brunak S, Staerfeldt HH, Ussery DW, “A DNA structural atlas for Escherichia coli”, The Journal of Molecular Biology, 299:907-930, (2000). https://doi.org/10.1006/jmbi.2000.3787 PubMed PMID: 10843847.
- Ussery D, Larsen TS, Wilkes KT, Friis C, Worning P, Krogh A, Brunak, “Genome organisation and chromatin structure in Escherichia coli”, Biochimie, 83:201-212, (2001). https://doi.org/10.1016/s0300-9084(00)01225-6 PubMed PMID: 11278070.
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR, “Genome sequence of enterohaemorrhagic Escherichia coli O157:H7”, Nature, 409:529-533, (2001). https://doi.org/10.1038/35054089 PubMed PMID: 11206551.
- Schembri MA, Ussery DW, Workman C, Hasman H, Klemm P, “DNA microarray analysis of fim mutations in Escherichia coli”, Molecular Genetics and Genomics, 267:721-729, (2002). Epub 20020621. https://doi.org/10.1007/s00438-002-0705-2 PubMed PMID: 12207220.
- Willenbrock H, Petersen A, Sekse C, Kiil K, Wasteson Y, Ussery DW, “Design of a seven-genome Escherichia coli microarray for comparative genomic profiling”, Journal of Bacteriology, 188:7713-7721, (2006). Epub 20060908. https://doi.org/10.1128/JB.01043-06 PubMed PMID: 16963574; PMCID: PMC1636325.
- Willenbrock H, Hallin PF, Wassenaar TM, Ussery DW, “Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray”, Genome Biology, 8:R267, (2007). https://doi.org/10.1186/gb-2007-8-12-r267 PubMed PMID: 18088402; PMCID: PMC2246269.
- Lukjancenko O, Wassenaar TM, Ussery DW,
“Comparison of 61 sequenced Escherichia coli genomes”, Microbial Ecology, 60:708-720, (2010). Epub 20100711. https://doi.org/10.1007/s00248-010-9717-3 PubMed PMID: 20623278; PMCID: PMC2974192. - Kaas RS, Friis C, Ussery DW, Aarestrup FM, “Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes”, BMC Genomics, 13:577, (2012). Epub 20121031. https://doi.org/10.1186/1471-2164-13-577 PubMed PMID: 23114024; PMCID: PMC3575317.
- Snipen LG, Ussery DW, “A domain sequence approach to pangenomics: applications to Escherichia coli”, F1000Research,1:19 (2012). Epub 20121001. https://doi.org/10.12688/f1000research.1-19.v2 PubMed PMID: 24555018; PMCID: PMC3901455.
- Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW, “Insights from 20 years of bacterial genome sequencing”, Functional & Integrative Genomics, 15:141-161, (2015). Epub 20150227. https://doi.org/10.1007/s10142-015-0433-4 PubMed PMID: 25722247; PMCID: PMC4361730.
- Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM, “Mash: fast genome and metagenome distance estimation using MinHash”, Genome Biology, 17:132, (2016). Epub 20160620. https://doi.org/10.1186/s13059-016-0997-x PubMed PMID: 27323842; PMCID: PMC4915045.
- Hunt M, Lima L, Shen W, Lees J, Iqbal Z, “AllTheBacteria – all bacterial genomes assembled, available and searchable”, BioRxiv, (2024). https://doi.org/10.1101/2024.03.08.584059
- Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36(6):1925-7. Epub 20191115. https://doi.org/10.1093/bioinformatics/btz848 PubMed PMID: 31730192; PMCID: PMC7703759.
- Parks DH, Chuvochina M, Reeves PR, Beatson SA, Hugenholtz P, Reclassification of Shigella species as later heterotypic synonyms of Escherichia coli in the Genome Taxonomy Database, BioRxiv, 2021; doi: https://doi.org/10.1101/2021.09.22.461432