This article explores the significance of G Protein-Coupled Receptors (GPCRs) in therapeutic drug development and strategies for advancing GPCR-targeted therapies, including the use of agonists, antagonists, biased agonism, and allosteric modulators
GPCRs: Key targets in therapeutic drug development for diverse diseases
G Protein-Coupled Receptors (GPCRs) are a large and diverse group of membrane proteins that play a crucial role in cellular communication and signal transduction. (1) Characterized by their seven transmembrane domains, GPCRs activate intracellular G proteins in response to extracellular signals such as hormones, neurotransmitters, and other signaling molecules. The GPCR superfamily consists of approximately 800 membrane proteins, sharing a common evolutionary origin and coupling primarily to intracellular transducers like G proteins and arrestins. These receptors are integral to complex signaling systems that modulate a wide array of physiological and pathophysiological processes. (2)
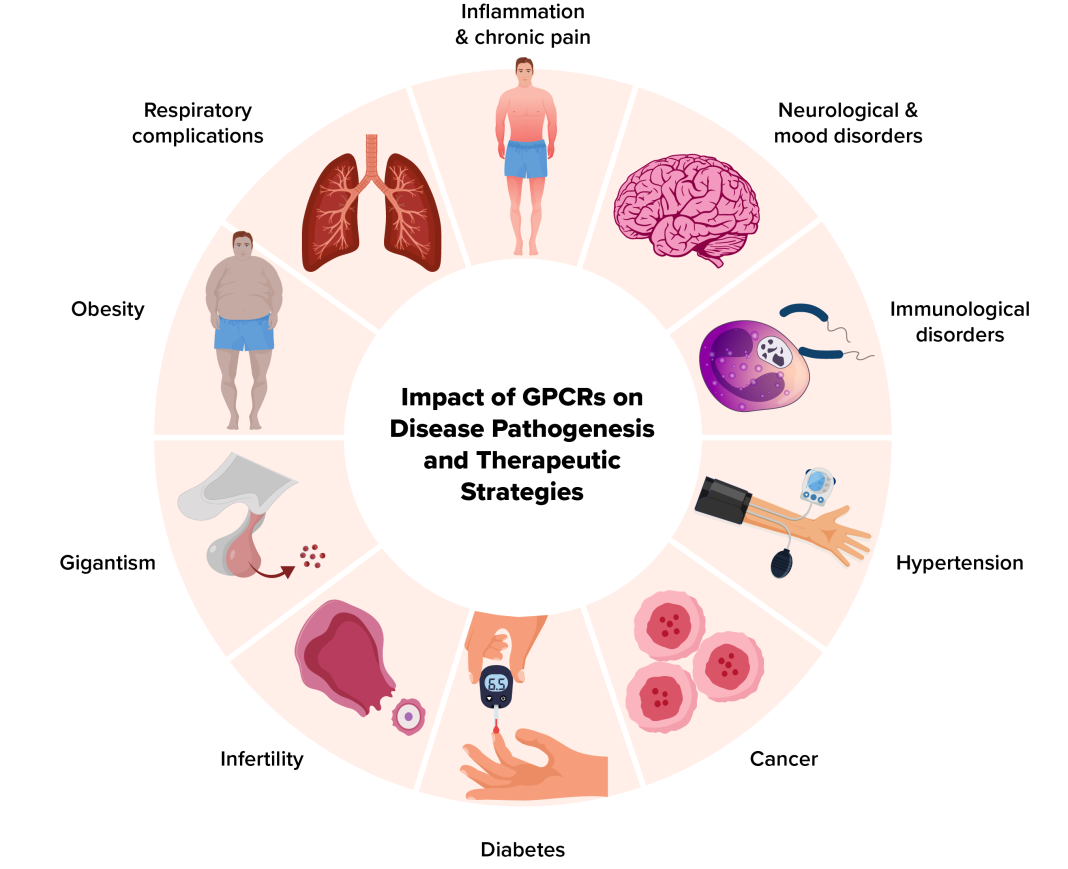
GPCRs are the target of roughly one-third of all therapeutic drugs, underscoring their significance in medicine. More than 500 distinct drugs targeting GPCRs for therapeutic effects are currently available in North America, Europe, Asia, and Australia. (3) These GPCR-targeted drugs account for approximately 36% of all approved drugs, reflecting the broad scope of GPCRs in mediating physiological processes and their therapeutic relevance, e.g. in inflammatory and autoimmune diseases, cancer, metabolic disorders, and central nervous system (CNS) disorders (Figure 1).
GPCRs are central regulators in the pathophysiology and treatment of inflammatory diseases
GPCRs are pivotal in inflammatory diseases like rheumatoid arthritis, IBD, psoriasis, and asthma, which involve chronic inflammation and tissue damage. These receptors regulate immune responses and inflammation, being central to these conditions’ pathophysiology. Present on immune cells such as T cells, B cells, macrophages, and neutrophils, GPCRs modulate cell trafficking, activation, and cytokine production: chemokine receptors, a GPCR subset, direct immune cells to inflammation sites. GPCRs also influence cytokine production, with certain activations leading to pro-inflammatory cytokines like TNF-α and IL-6. In tissues like the gut and skin, GPCRs maintain barrier integrity, and their signaling disruption can worsen inflammation, as seen in IBD – some GPCRs aid in resolving inflammation by clearing inflammatory cells and restoring tissue homeostasis.
Advancing GPCR-targeted therapies: Strategies and challenges in treating inflammatory diseases
Therapeutic targeting of GPCRs is promising for inflammatory diseases due to their central role in inflammation. Strategies include using agonists and antagonists to modulate GPCR activity, such as CCR5 antagonists to reduce inflammation. Biased agonism involves designing ligands that selectively activate beneficial pathways, minimizing side effects. Allosteric modulators bind to distinct sites, allowing fine-tuning of GPCR activity without competing with natural ligands.
Challenges include the complexity of GPCR signaling, potential desensitization, and functional overlap, requiring precise targeting to avoid side effects. Future research aims to better understand GPCR roles in inflammation and develop more selective modulators. Advances in structural biology and computational modeling are aiding the design of novel therapies, with improved compound synthesis needed for future GPCR-based treatments.
Accelerating drug discovery: Leveraging automation and Machine Learning for rapid GPCR drug development
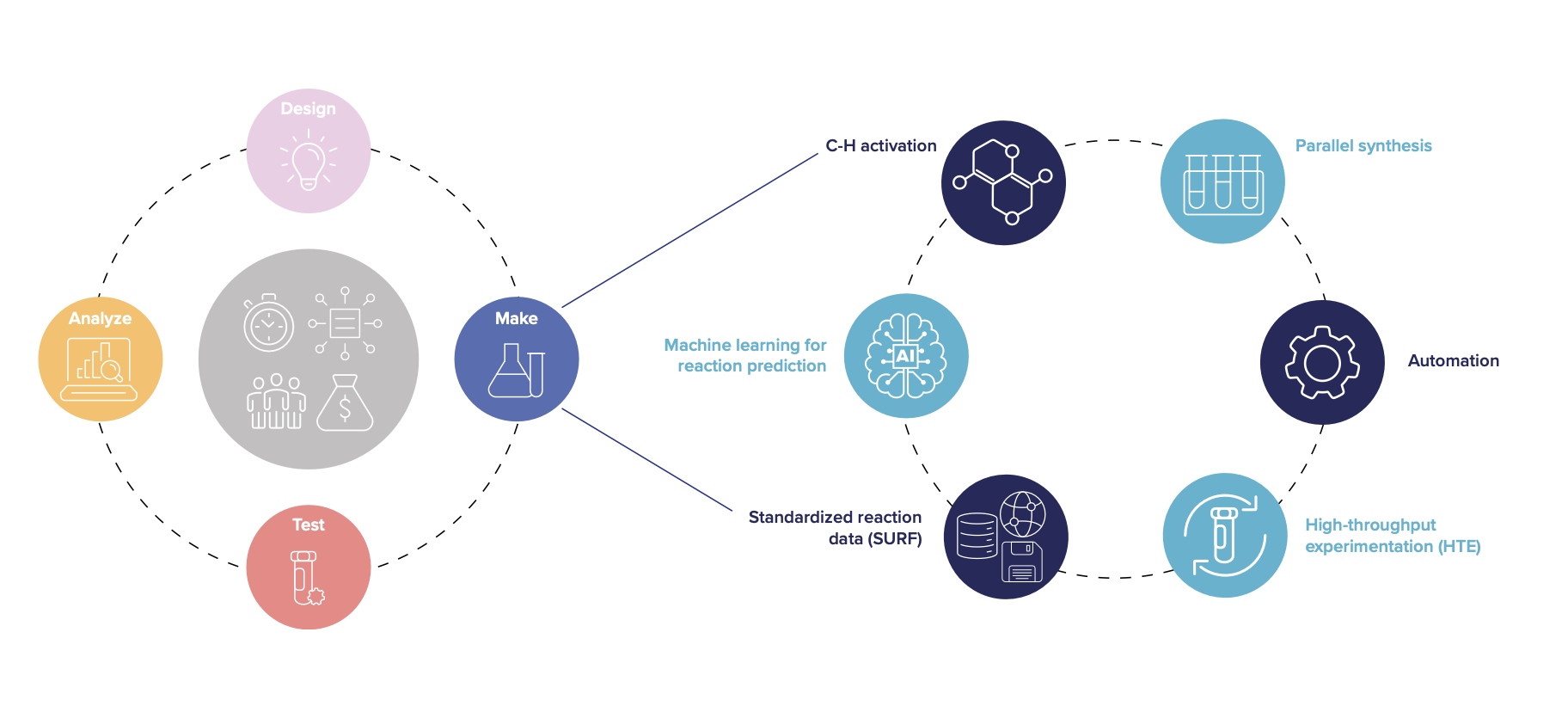
Automation, data, and Machine Learning significantly enhance the success of selective C-H activation reactions by addressing their inherent complexity. (4) High-throughput experimentation (HTE) enables simultaneous reactions, improving synthesis planning and success rates. (5) Machine Learning tools, reliant on high-quality data, have effectively addressed challenges in reaction prediction and synthesis planning. Roche’s Simple User-Friendly Reaction Format (SURF) facilitates comprehensive reporting of reaction data. (6)
The Design-Make-Test-Analyze (DMTA) cycle, combined with advanced methodologies, accelerates the hit-to-lead optimization phase in drug discovery (Figure 2). Integrating reaction prediction with other predictive tools in computer-aided drug design expedites small molecule development. A proof- of-concept study demonstrated accelerated hit-to-lead optimization, applicable to other drug discovery projects. (7) Enhancing the synthesis step in the DMTA cycle can significantly impact development timelines for new medicines.
These innovative methodologies promise faster generation of GPCR drugs to address inflammatory diseases with high unmet medical needs. (8)
References
- Hauser AS, Attwood MM et al. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017 Dec;16(12):829-842. doi: https://doi.org/10.1038/nrd.2017.178.
- Jobe A, Vijayan R. Orphan G protein-coupled receptors: the ongoing search for a home. Front Pharmacol. 2024 Feb 29;15:1349097. doi: https://doi.org/10.3389/fphar.2024.1349097.
- Lorente JS, Sokolov AV et al. GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2025 Mar 3. doi: https://doi.org/10.1038/s41573-025-01139-y.
- a) Cernak T, Dykstra KD et al. The medicinal chemist’s toolbox for late stage functionalization of drug-like molecules. Chem Soc Rev. 2016 Feb 7;45(3):546-76. doi: 10.1039/c5cs00628g; b) Nippa DF, Atz K et al. Identifying opportunities for late-stage C-H alkylation with high-throughput experimentation and in silico reaction screening. Commun Chem. 2023 Nov 20;6(1):256. doi: https://doi.org/10.1038/s42004-023-01047-5.
- Nippa DF, Atz K et al. Enabling late-stage drug diversification by high-throughput experimentation with geometric deep learning. Nat Chem. 2024 Feb;16(2):239- 248. doi: https://doi.org/10.1038/s41557-023-01360-5.
- Nippa DF, Müller AT et al. Simple User-Friendly Reaction Format. Mol Inform. 2025 Jan;44(1):e202400361. doi: https://doi.org/10.1002/minf.202400361.
- Nippa DF, Atz K et al. Expediting hit-to-lead progression in drug discovery through reaction prediction and multi-objective molecular optimization. ChemRxiv. 2025; doi:https://doi.org/10.26434/chemrxiv-2025-0lxhw
- a) Julie Blaising and Philip Smith (2024), “Therapeutic potential of the endocannabinoid system to treat chronic pain in inflammatory disease”, Open Access Government April 2024, pp.158-159. Available at https://www.openaccessgovernment.org/article/therapeutic-potential-of-the-endocannabinoid-system-to-treat-chronic-pain-in-inflammatory-disease/174807/. (Accessed: 03 Mar 2025); b) Julie Blaising (2024), “Protein kinases for combating inflammatory disease”, Open Access Government July 2024, pp.150-151. Available at https://www.openaccessgovernment.org/article/protein-kinases-for-combating-inflammatory-disease/178201/. (Accessed: 09 Mar 2025); c) Uwe Grether and Julie Blaising (2024), “MAGL inhibition:
A novel treatment option for combating inflammatory disease?”, Open Access Government January 2024, pp.48-49. Available at https://www.openaccessgovernment.org/article/magl-inhibition-a-novel-treatment-option-for-combating-inflammatory-disease/172631/. (Accessed: 09 Mar 2025).