Professor Jatinder Lamba from the Department of Pharmacotherapy and Translational Research gives an in-depth perspective on acute myeloid leukaemia (AML) and cytarabine pharmacogenomics
Acute myeloid leukaemia (AML) is the cancer of the myeloid precursor stem cells characterised by the appearance of immature, abnormal myeloid cells in bone marrow and other organs. AML is a very heterogeneous disease with various subtypes classified based on the morphology, immnuophenotype and cytogenetics.
Despite advances in recent years, the 5-year survival rates for AML are ~68% for children younger than 15 years and ~57% for children between 15-19 years. The utilisation of multiple clinical, cytogenetic and other molecular features that are associated with the response has helped in identification of patient’s more/less likely to respond.
Additionally, minimal residual disease (MRD) after induction 1 has been identified as a powerful predictor of poor outcome. The nucleoside analog, cytarabine (ara-C), has been the mainstay of AML chemotherapy for more than 40 years1,2. The most common treatment strategy includes induction therapy, which consists of 3+7 regimen with daunorubicin (45 mg/m2 per day for 3 days) and cytarabine 100mg/m2 for 7 days). This induction regimen has been shown to induce remission in 60-70% of patients. Cytarabine, in combination with varying doses of anthracycline and addition of a third agent (cladribine, clofarabine and more recently CD33 targeted therapies such as gemtuzumab ozogamicin etc.), has been the standard of practice for a few years now.
However, despite the major advances in AML drug combination strategies and treatment options, resistance and disease remission represent our major obstacle to reach the desired outcomes3,4. Although a significant proportion of patients achieve complete remission after initial therapy, most of these experience relapse which has a significant impact on long-term survival rates5
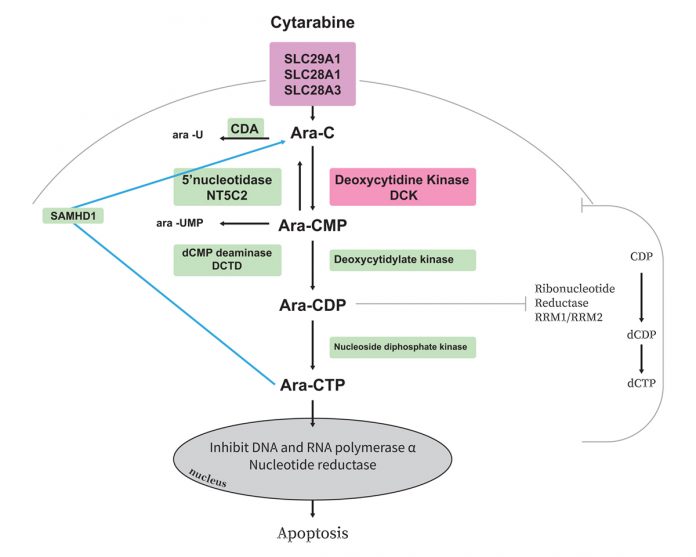
One of the most common and severe toxicities associated with cytarabine is myelosuppression. Cytarabine is a prodrug requiring activation to ara-CTP through series of phosphorylation steps and this triphosphate form of ara-C inhibits DNA polymerase-α and β and competes, with deoxycytidine triphosphate (dCTP) for incorporation into DNA and RNA thereby inhibiting DNA synthesis and triggering leukaemic cells6-8. Thus, intracellular levels of the active ara-CTP are critical for achieving significant leukaemic cell death. Previous in vitro studies have shown that the intracellular concentrations of ara-CTP are higher in ara-C sensitive cells, than in resistant cells.
Further, leukaemic cells from patients with chronic myelogenous leukaemia (which is not responsive to ara-C), have only half the ara- CTP levels, as compared to leukaemic cells from patients with AML (which is responsive to ara-C)4. Thus, one of the mechanism underlying ara-C resistance is insufficient intracellular levels of the active triphosphate metabolite ara-CTP, which may be due to:
- Inefficient cellular uptake due to low levels of the transporters (SLC29A1, SLC28A3 and SLC28A1);
- Reduced activation due to alterations in enzymes as DCK, CMPK1;
- Increased inactivation rates due to NT5C2, CDA, or DCTD and more recently identified enzyme SAMHD19; and/or
- Increased cellular dCTP pools, that can compete with DNA incorporation of ara-CTP and inhibit DCK activity which, in turn, are regulated by the enzyme ribonucleotide reductase (consisting of RRM1 and RRM2 subunits). Figure shows metabolic pathway and thus key players that might impact cytarabine activation and thus its therapeutic benefit.
Single nucleotide polymorphisms (SNPs) in genes involved in transport, activation and inactivation of cytarabine can influence the intracellular ara-CTP levels by influencing the expression and activity of these genes and this, in turn, can also influence the clinical outcome of the patients treated with ara-C.
Several studies so far have investigated some of the key players and have shown an association of SNPs within key candidate cytarabine metabolism genes and clinical response in AML patients, or with cytarabine in vitro chemosensitvity10. Our group has previously reported an association of SNPs in DCK, NT5C2 and RRMs with intracellular levels of ara-CTP in leukaemic cells, as well as clinical response to AML11-15.
Despite these efforts, one of the biggest gaps in the literature is that most of the previous studies have focused on selective genes and so far, no study has performed a comprehensive and integrated analysis of genetic variation in the complete cytarabine metabolic pathway. It is important to note that simultaneous contribution of multiple genes can impact the drug activation and thus response, so it is very critical to the field of pharmacogenomics around AML/paediatric and AML is still in its infancy.
In summary, AML is a very heterogeneous disease, with multiple subgroups that impact the therapeutic outcome. Pharmacogenomics has the potential to improve the clinical outcome in AML, although there are limited studies, results so far indicate the potential for SNPs in activating, inactivating enzymes, as well as drug transporter to be of clinical relevance. The integration of pharmacogenetic markers with prognostic markers in larger clinical cohorts can advance our ability to designed personalised therapy for patients to achieve greatest therapeutic benefit.
Funding is provided by National Cancer Institute through grant R01-CA132946 (LAMBA NAD POUNDS) to support the work on AML Pharmacogenomics.
- Braess, J. [Acute myeloid Leukemia]. Dtsch Med Wochenschr 141, 1748-1751 (2016).
- Short, N.J. & Ravandi, F. Acute Myeloid Leukemia: Past, Present, and Prospects for the Future. Clin Lymphoma Myeloma Leuk 16 Suppl, S25-9 (2016).
- Rowe, J.M. AML in 2016: Where we are now? Best Pract Res Clin Haematol 29, 315-319 (2016).
- Abraham, A. et al. RNA expression of genes involved in cytarabine metabolism and transport predicts cytarabine response in acute myeloid leukemia. Pharmacogenomics 16, 877-90 (2015).
- Ravandi, F. Relapsed acute myeloid leukemia: why is there no standard of care? Best Pract Res Clin Haematol 26, 253-9 (2013).
- Major, P.P., Egan, E.M., Beardsley, G.P., Minden, M.D. & Kufe, D.W. Lethality of human myeloblasts correlates with the incorporation of arabinofuranosylcytosine into DNA. Proc Natl Acad Sci U S A 78, 3235-9 (1981).
- Major, P.P., Sargent, L., Egan, E.M. & Kufe, D.W. Correlation of thymidine-enhanced incorporation of ara-C in deoxyribonucleic acid with increased cell kill. Biochem Pharmacol 30, 2221-4 (1981).
- Yates, J. et al. Cytosine arabinoside with daunorubicin or adriamycin for therapy of acute myelocytic leukemia: a CALGB study. Blood 60, 454-62 (1982).
- Schneider, C. et al. SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat Med 23, 250-255 (2017).
- Bhise N., C.L., and Lamba J.K. Genome-Wide Association Studies – From polymorphisms to Personalized Medicine, Chapter 19: Pharmacogenetic studies in pediatric acute myeloid leukemia, (Cambridge University Press, 2016).
- Cao, X. et al. RRM1 and RRM2 pharmacogenetics: association with phenotypes in HapMap cell lines and acute myeloid leukemia patients. Pharmacogenomics 14, 1449-66 (2013).
- Lamba, J.K. Pharmacogenomics of cytarabine in childhood leukemia. Pharmacogenomics 12, 1629-32 (2011).
- Mitra, A.K. et al. Genetic variants in cytosolic 5’-nucleotidase II are associated with its expression and cytarabine sensitivity in HapMap cell lines and in patients with acute myeloid leukemia. J Pharmacol Exp Ther 339, 9-23 (2011).
- Lamba, J.K. Genetic factors influencing cytarabine therapy.
Pharmacogenomics 10, 1657-74 (2009). - Lamba, J.K. et al. Pharmacogenetics of deoxycytidine kinase: identification and characterization of novel genetic variants.
J Pharmacol Exp Ther 323, 935-45 (2007).
Lamba Lab members involved in the cytarabine pharmacogenomics research include AbdelRehman Elsayed, Dr. Roya Rafiee and Salma Bargal
Please note: this is a commercial profile
Professor Jatinder Lamba
Department of Pharmacotherapy and
Translational Research
Tel: +1 352 273 6425