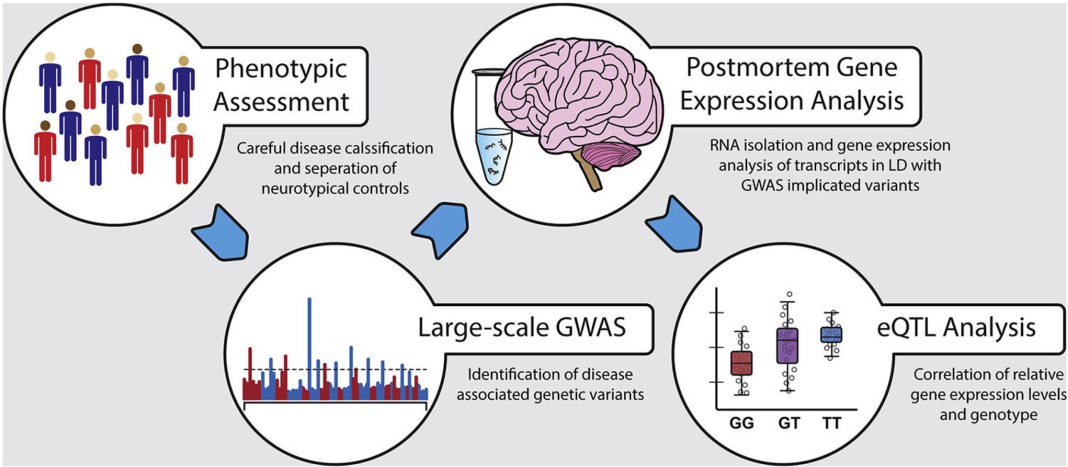
Vladimir Vladimirov discusses two key advancements in genome-wide association studies (GWAS) and gene expression data to better understand the biological basis of mental disorders
The economic burden of severe mental disorders, such as schizophrenia (SZ), bipolar disorder (PB), or major depressive disorder (MDD), is estimated at approximately $280bn annually. Most importantly, these are chronic and currently uncurable disorders with significant negative impacts on society and families. Even though we are already a quarter of a century into the 21st century, the currently available medication for the treatment of these disorders has not improved since the early 80s. A primary reason for the lack of novel and potentially more effective (with fewer side effects) drugs for the treatment of mental disorders is that we did not have a good grasp of the biological and genetic mechanisms underlying the disease etiology. However, our understanding of the genetics, if not yet biological, basis of psychiatric disorders has substantially improved over the last two decades, notably due to significant technological breakthroughs. Below, I give a short overview of two critical advancements in the field related to the integration of genome-wide associations scans (GWAS) and transcriptome (gene expression) data generated in psychiatric patients as one of the pathways of moving forward toward elucidating the biological basis of mental disorders.
Understanding the genetic variants associated with SZ, BP, and MDD
In the past decade, a massive and concerted effort from multiple research groups has led to the generation of large ‘mega’ GWAS, analyzing millions of genotyped and imputed single nucleotide polymorphisms (SNP) generated from thousands of subjects. It is the scale of these studies that have enabled the first detection of robust and replicable genetic variants associated with SZ, BP, and MDD. However, a significant challenge of the post-GWAS era is understanding how these clinically relevant genetic findings will lead to identifying molecular processes underlying disease etiopathology. The main reason for this is that except for some common disorders, such as colon cancer or diabetes, very little is known about the biological mechanism(s) by which the genetic variants contribute to complex behavioral psychopathologies. The increased success in identifying the molecular pathways contributing to cancer or diabetes has also been due to our ability to study these processes in the etiologically relevant tissues.
The human brain is the etiologically relevant tissue for studying molecular processes involved in neuropsychiatric disorders. However, collecting postmortem brain tissue is difficult due to extensive logistic requirements and substantial ethical and cultural hurdles. Regardless, efforts to collect postmortem brains from subjects with neuropsychiatric disorders and controls have been undergoing for over three decades. Research centers, including (but not limited to) the Lieber Institute for Brain Development (LIBD), the CommondMind Consortium (CMC), and Stanley Medical Research Institute (SMRI), have amassed thousands of brains with data on various neuropsychiatric phenotypes.
We have already alluded to a well-recognized limitation of GWAS data: their inability to elucidate the biological mechanisms involving the identified associated variants. Thus, the first step in understanding the biological function of implicated SNPs consists in assessing its impact on gene expression, i.e., essentially identifying expression quantitative trait loci (eQTLs) (Figure 1), since gene expression is considered the most intermediate phenotype between genetic polymorphisms and downstream biological processes. Although nearly all variants identified by GWAS occur outside of protein-coding genomic regions, these variants can still regulate gene expression indirectly through various means. For example, the associated polymorphisms can generally affect gene expression or target transcript gene isoforms via SNPs, impacting alternative splicing mechanisms. Another mechanism by which significant GWAS polymorphisms affect gene expression or specific transcript abundance is by interfering with miRNA function, i.e., influencing the interactions between miRNAs and their gene targets. Such effects have already been observed in a limited number of studies investigating neuropsychiatric disorders such as MDD, SCZ, and BPD.
Additionally, sequence variants have been known to modify sequence-specific methylation patterning at gene regulatory elements, effectively altering gene expression. Thus, determining the impact of genetic variants on gene regulation is crucial for elucidating the mechanisms by which variants increase the risk of disease.
Human post-mortem brain research challenges
However, to properly use postmortem brain tissue to study neuropsychiatric disorders, it is paramount that we use brain tissues from continuously increasing sample sizes and, perhaps, even more critically, collect brains from ethnically diverse populations. While the establishment of various large postmortem brain collection centers has begun to address the need for greater sample sizes, expanding the ethnic diversity of potential donors remains a necessity for providing insight into the population-specific disease-associated genetic mechanisms. For instance, virtually all available postmortem brain banks worldwide are composed primarily of subjects with European or North American genetic backgrounds. The importance of integrating GWAS and transcriptome data from the same population can be further surmised from the GWAS data alone, which have long identified population stratification as a major confounding factor in determining true disease association signals due to variation in allele frequencies between ethnic groups. Disregarding population-specific genetic variation can lead to substantial difficulties in interpreting the eQTL data, especially for SNPs with significantly varying allele frequencies between populations. The limited tissue contribution to postmortem brain banks by donors from different ethnic groups may be a result of negative social, political, and cultural ideations concerning organ donation. It is well understood, at least in the North American medical community, that minority groups are less likely to participate in organ donation compared to Caucasians. For these reasons, initiating international collaborations to establish postmortem brain repositories dedicated to expanding the ethnic diversity of potential donors would greatly improve research outcomes for understanding how genetic predispositions impact neuropsychiatric etiopathology across different populations.
In conclusion, integrating findings from large GWAS and high-throughput gene expression studies will inform the functional relevance of disease- associated risk variants. While in recent years, the statistical power has become less concerning, the lack of ethnic diversity among the current postmortem brain banks still significantly impedes the progress toward understanding the genetic mechanisms underlying neuropsychiatric disorders. Within the scientific community, a greater emphasis should be placed on developing robust, ethnically diverse postmortem brain databases through public education on the importance of organ donation and international scientific collaboration. The wealth of information from expanded postmortem brain studies will increase our understanding of individual differences’ roles in disease susceptibility and yield important insights into ethnicity-specific occurrence rates, progression, and treatment of neuropsychiatric disorders.