Alan Herbert, Founder and President of InsideOutBio, discusses alternative DNA conformations and understanding of their biological functions
One of the great things about science is that every time you think you really understand things and can’t imagine how they could work differently, life surprises you. The biggest mistake you can make is believing that nature works according to your best design. Take physics, for example – first, there were the four humors, then atoms, then protons and electrons, then even more particles, and then quarks – who knows what’s next? Similarly, biology is undergoing its own progression, and we are finding that there is more to life than just Watson and Crick’s DNA.
Interestingly, about the time the Watson and Crick model of B-DNA was described, it was already known that DNA can adopt different structures. The model of B-DNA was highly predictive and enabled a revolution that culminated in the advances of genetic engineering that led to the modern-day alchemy of being able to precisely edit the genome in various therapeutic ways. It is easy to think that we were done. Of course, you are only as good as your methods and the data representing your worldview. But when trying to understand something as complex as life and how it evolved, it is natural to try and simplify the problem – the complementary base pairing of the double helix provides an easy way to copy DNA. The DNA copy can then transmit genetic information from one generation to the next.
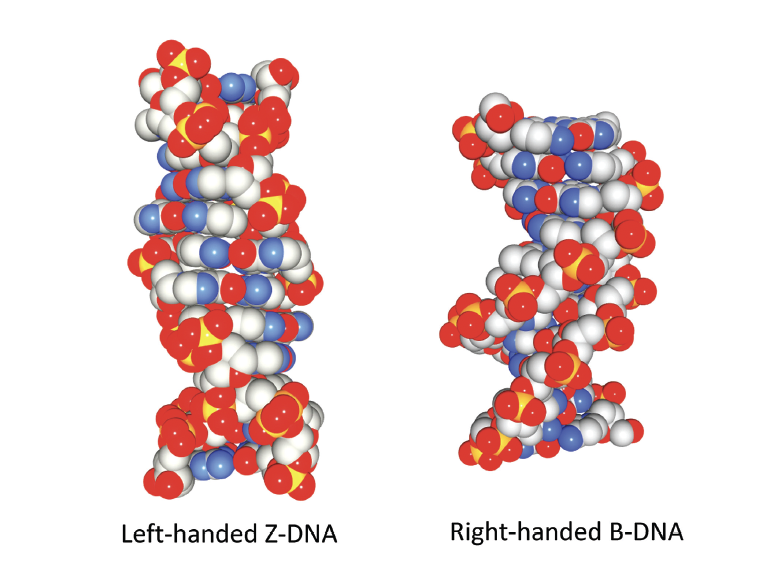
But we are not even close to knowing what else DNA does in a cell. I consider some of the questions set aside in my book, Flipons: The Discovery of Z-DNA and Soft-Wired Genomes (CRC Press, 2024). The story of alternative DNA conformations focuses on higher energy DNA structures formed dynamically within the cell. In particular, the story of the discovery of left-handed DNA and its biological functions is related. Z-DNA helix is wrapped to the left, unlike the Watson and Crick DNA that twists to the right. The existence of Z-DNA was proven somewhat surprisingly, being detected in the first DNA crystal ever made. However, the odd form of DNA, although beautiful to the eye, did not appear to explain anything biologically. In reality, the methods did not exist to answer the question of ‘what does Z-DNA do, if anything?’

Zα protein domain
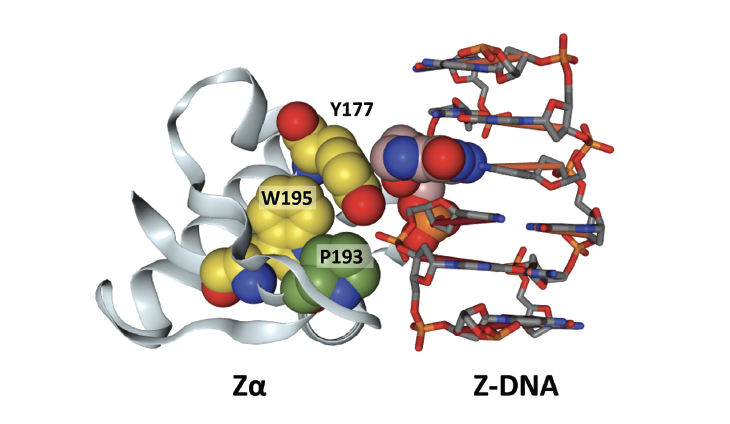
Since then, new methods have led to the discovery of the Zα protein domain that binds with high affinity to Z-DNA. Now, through a series of biochemical, genetic, and cell biology studies, we have answers to how these alternative DNAs change the readout of information from our genes. While the genome contains highly diverse sequences that code for specific protein functions, the alternative DNA folds are often formed by elements in which the bases have distinct repeat patterns. The repeats have low information content since many of each type exist throughout the genome. Indeed, over 50% of the genome contains repeats of one form or another. The change in flipon conformation is highly informative. It requires energy and flags an active gene. The flip from B-DNA to another DNA structure alters the RNA messages read from the gene: trade of energy for information.
The repeat elements that adopt the various possible alternative conformations under physiological conditions are called flipons. They act as binary elements, being one structure or the other. They enable the assembly of conformation-specific complexes whose function depends on which form of DNA is bound. Some complexes can increase gene expression, while others suppress the readout from that region. The alternative conformations can also change the processing of RNA transcripts. By changing how the RNA messengers are spliced and edited, a variety of proteins can be produced from a single gene by the ribosomal translation machinery. The book describes these discoveries both in DNA structure and the logic behind these binary switches.
Interestingly, the Zα domain can recognise Z-DNA and Z-RNA (collectively ZNA). Generally, both form transiently in cells and are present at low amounts. However, when cells become dysregulated or infected by viruses, the levels of ZNA can increase dramatically. That event triggers one of the two proteins in the human genome with a Zα domain. One protein ZBP1 senses the formation of ZNAs and can amplify immune responses. In some cases, a highly inflammatory form of cell death is initiated that ramps up immune responses against the threat. The other Zα domain protein ADAR1 negatively regulates these inflammatory responses against self-RNAs, dampening down the damage to bystander cells. Some viruses, such as smallpox, try to interfere with these pathways by encoding their own version of a Zα domain. Interestingly, the host often produces the Z-RNA that drives the response, not viral flipons.
The flipons are part of the genome that theRNApolymerizingmachinerydoes not normally transcribe. However, if something goes wrong, then these flipons are read out. When the host makes the ZNA they encode, the Zα domain proteins tell the cell that it is under attack. Because these flipons enhance survival against threats, they are subject to natural selection. The cell can defend against known or unknown viruses using this built-in alarm system.
Interestingly, the Zα domain traces back to the earliest multicellular organisms but was lost in the organisms used for classical genetic studies. Instead, bacteria, worms, flies, and plants use a different mechanism to protect against threats –based on RNA-directed suppression of RNAs and DNAs produced by pathogens. No wonder the first generation of molecular biologists found no evidence for Z-DNA or Z-RNA when they studied these systems.
Intriguingly, the Zα domain has itself evolved. While it retains the α-helix that specifically recognizes ZNAs, it has changed the β-sheet wing by extending its length and changing the residues that interact with DNA. Of great interest is that the modified domain is associated with transcription. The result is most clearly seen in the third domain of life: the archaea. Tracking forward, these Zα- related proteins form complexes with RNA polymerases. This suggests that Z-DNA is associated with the readout of RNA from genes. Indeed, the finding explains why there is a concentration in the region of genes of sequences with a propensity to form Z-DNA that promote transcription.
The book further describes how the conformation of flipons can be regulated by small RNAs to change the readout of genetic information. By targeting the flipon binary switch, the small RNAs can change the complexes assembled at promoters. The use of such RNAs to control gene expression in other ways is also described. Examples include those RNAs that guide the editing and splicing of RNA transcripts as they are made. In some organisms, the RNAs restore defective genomes to make the proteins required to survive. These different RNA-guided systems underlie the development of a new class of therapeutic RNAs just entering the clinic. They offer the ability to reverse genetic disease and to alter cellular pathways at the level of RNA without modifying genomic DNA, overcoming some of the objections to irreversibly altering the human genome using CRISPR or Fanzor technologies.

Overall, the book uses the discovery of Z-DNA as an example to show how the scientific process works. The narrative describes the initial hype followed by a valley where there was no funding or jobs for those who worked on something deemed ‘irrelevant.’ The pattern follows what has occurred many times over the history of scientific advances. While we consider ourselves sophisticated and immune to past mistakes, we certainly are not. Many historical parallels are presented, not by a philosopher but by someone at the frontier of change. The topics discussed include the resistance of the old to the new, the tensions between junior and senior scientists, both male and female, the misattribution of discoveries, and the role of critics and journal editors in the process. While the discovery of B-DNA has led us to many advances, alternative DNA conformations will help us better understand the logic of how cells work and the evolution of multicellular organisms.
Flipons: The Discovery of Z-DNA and Soft-Wired Genomes (CRC Press, 2024)

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.