Charles W. Carter, Jr, from the Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill explores the roots of genetic readout in the inherent structural duality of DNA and how genetic coding expanded its potential, enabling life to emerge
One of Nature’s most ingenious inventions, the genetic code unlocked a vast molecular machinery enabling independently living cells to emerge from the complex chemistry that preceded the origin of life.
Genetic Coding: Nature’s Operating System
Smaller, more diverse amino acid subunits packing around protein active sites forged a billion-fold amplification over the chemical engineering possible using only four larger nucleotide bases. That amplification, translation, depends on catalyzed assignment of each of the 20 amino acids in proteins to one or more of the 64 symbols – three sequential nucleotide bases called codons – constructed using the letters A, T, C, and G. Genetic coding provided a programming language to write symbolic programs (genes). It is thus Nature’s operating system (OS).
How coding interprets genes became evident soon after Watson and Crick revealed that recognition of A by T (DNA) or U (RNA) and of G by C held the two antiparallel strands of the double helix together. It nonetheless even now remains mysterious to us, the only descendants able to ask the question, how Nature created its OS from scratch, with neither programs, alphabets, nor symbols. It is the quintessential challenge posed by the origin of life.
That challenge hides a crucial, rarely stated question. The molecular assignment catalysts—aminoacyl-tRNA synthetases (aaRS)—are made from genes written in the very language they must implement; they are reflexive (1). Roots of that reflexivity lie deep in evolutionary molecular biology—a historical progression in which each step exploits what was already available, while enabling the next.
Was Nature’s boot block a bidirectional gene?
Pursuing the operating system metaphor, my colleagues and I are searching for Nature’s “boot-block” – a necessary and sufficient instruction set to elaborate the full coding table and the genes to implement it.
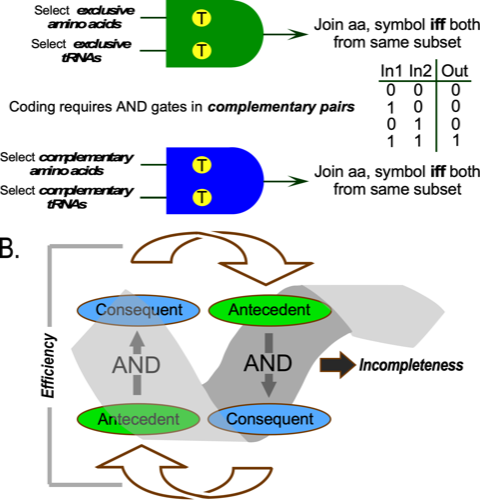
AARS work like computer AND gates, selecting from amino acid and tRNA substrates and forming a covalent bond between them if and only if both selections are correct (Fig. 1A). Discovery that aaRS come in two parallel, distinct structural families differentiated the amino acids naturally into Classes, I and II(2). Much experimental data now support the proposal(3,4) that Class I and II aaRS were originally encoded on opposite strands of the same ancestral gene. Translation products – called “protozymes” – from both strands of a designed bidirectional gene encoding just the 46-residue ATP and amino acid binding sites for the two aaRS Classes(5) speed up amino acid activation a million fold, overcoming the slowest step in protein synthesis.
Amino acid size and polarity differentiate the canonical amino acids(6) and determine how proteins fold(7). Remarkably, Nature chose codons that pass base-pairing complementarity from the gene on into the proteome. Discrimination by Class I and II aaRS of large vs small amino acids, and tRNA acceptor stem binding from the major vs the minor groove are direct consequences of bidirectional coding (8- 10). The details allowed us to elaborate the earliest coding by primordial aaRS and tRNAs(11).
A one-bit coding alphabet for two contrasting types of amino acids implemented by two, mutually exclusive, aaRS•tRNA “cognate pairs” is certainly necessary, but may or may not be sufficient for this purpose. Nevertheless, a bidirectional ancestral aaRS gene serves as an excellent, experimentally testable model for booting Nature’s OS.
Ancestral aaRS launched catalysis, genetics, and bioenergetics
The properties – catalysis and amino acid activation – of the bidirectional protozyme gene(5) suggest that it contained seeds for elaborating the proteome. The Class I protozyme appears, with variation, in nearly a third of known proteins(12). They all share an amino acid packing motif that serves as a conformational master switch governing domain motion and the efficient conversion of ATP hydrolysis free energy in one Class I aaRS(13,14). That example exhibits allosteric behavior shared by many contemporary mechanical and signaling enzymes, all arguably genetic descendants of the protozyme(15)
Understanding contemporary aaRS genes: Where to go from here?
Deconstructing the patchwork of contemporary aaRS genes, we’ve characterized a nested hierarchy of functional modules from both aaRS classes as experimental models(16-20) to explore these challenges:
“Urzymes” are 130-residue excerpts that subsume and have a thousand times greater catalytic proficiency than protozymes(5). Urzymes recognize and acylate cognate tRNAs. However, although sequence data(21) suggest how to construct a bidirectional urzyme gene, we have not yet succeeded. To do so will require inspired use of phylogenetic methods to reconstruct the earliest proteome(22).
Are reconstructed urzyme•tRNA cognate pairs specific enough to suggest plausible, experimentally testable pathways to the remaining aaRS?
Catalysis, genetics, and bioenergetics all demonstrate that Nature uses reciprocally coupled gating(21) (Fig. 1B) to filter optimal behaviors from large related, but less active populations. AND gates allow passage if and only if a certain condition is met. Coupling the antecedent of one AND gate head-to-tail to the consequent of another creates a powerful filter bypassing Darwinian natural selection. Could reciprocally coupled gating have coordinated the self-organization of all three phenomena?
References
- Carter, C. W., Jr & Wills, P. R. MBE 35, 269-286, (2018).
- Eriani, G., et. al., Nature 347, 203-206 (1990).
- Rodin, S. N. & Ohno, S. Orig. Life Evol. Biosph. 25, 565-589 (1995).
- Carter, C. W., Jr. & Wills, P. R. Ann. Rev. Biochem. 90, 349- 373 (2021).
- Martinez, L. et al. J. Biol. Chem. 290, 19710–1972 (2015).
- Carter, C. W., Jr. & Wolfenden, R. Proc. Nat. Acad. Sci. USA 112 7489-7494 (2015).
- Wolfenden, R., et. al., Proc. Nat. Acad. Sci. USA 112 7484- 7488 (2015).
- Carter, C. W., Jr & Wills, P. R. NAR 46, 9667–9683 (2018).
- Carter, C. W., Jr & Wills, P. R. IUBMB Life 71, 1088–1098 (2019).
- Carter, C. W., Jr. & Wills, P. R. BioSystems 183 103979 (2019).
- Schimmel, P., et. al., PNA. USA 90, 8763-8768 (1993).
- Cammer, S. & Carter, C. W., Jr. Bioinformatics 26, 709- 714 (2010).
- Carter, C. W., Jr. Ann.l Rev. Biophys. 46, 433-453 (2017).
- Weinreb, V., Li, L. & Carter, C. W., Jr. A Structure 20, 128- 138 (2012).
- Carter, C. W., Jr. Proteins: Struct., Funct., Bioinf. 88, 710– 717 (2019).
- Carter, C. W., Jr. JBC. 289, 30213–30220 (2014).
- Li, L., Francklyn, C. & Carter, C. W., Jr. JBC 288, 26856- 26863 (2013).
- Li, L., et. al., JBC 286, 10387-10395 (2011).
- Pham, Y. et al. JBC 285, 38590-38601 (2010).
- Pham, Y. et al. Mol. Cell 25, 851-862 (2007).
- Chandrasekaran, S. N., MBE 30, 1588-1604 (2013).
- Carter, C. W., Jr., et. al., IJMS 23, 1520 (2022).
- Carter, C. W., Jr. & Wills, P. R. Biomolecules 11, 265 (2021).

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.


