Host defences operate to prevent ‘ancient viruses’ from ever jumping but, in cancers, cells lose multiple layers of ‘epigenetic’ control, and this can lead to the awakening of jumping or ‘retrotransposition’ of ancient viruses
The ‘dark genome’ refers to the extensive amount of uncharacterized DNA within the human genome and contains strings of repetitive DNA sequences. Most of the dark genome is derived from transposable elements, which are ancient genome invaders, a minority of which can still replicate and jump to invade new genomic locations. Host defences operate to prevent these transposable elements or ‘ancient viruses’ from ever jumping. Still, in cancers, cells lose multiple layers of ‘epigenetic’ control and this can lead to the awakening of jumping or ‘retrotransposition’ of ancient viruses. This jumping into our genes can disrupt their function and cause disease.
How can research aimed at studying the function of the dark genome benefit us?
Cutting-edge research in this area will shed light on 1. Novel mutations that are linked to diseases such as cancer and which could be targeted as therapies, 2. Which parts of our genome could be used in cancer vaccines to direct immune responses to clear the tumour, and 3. How the dark genome contributes to the evolution of cancer.
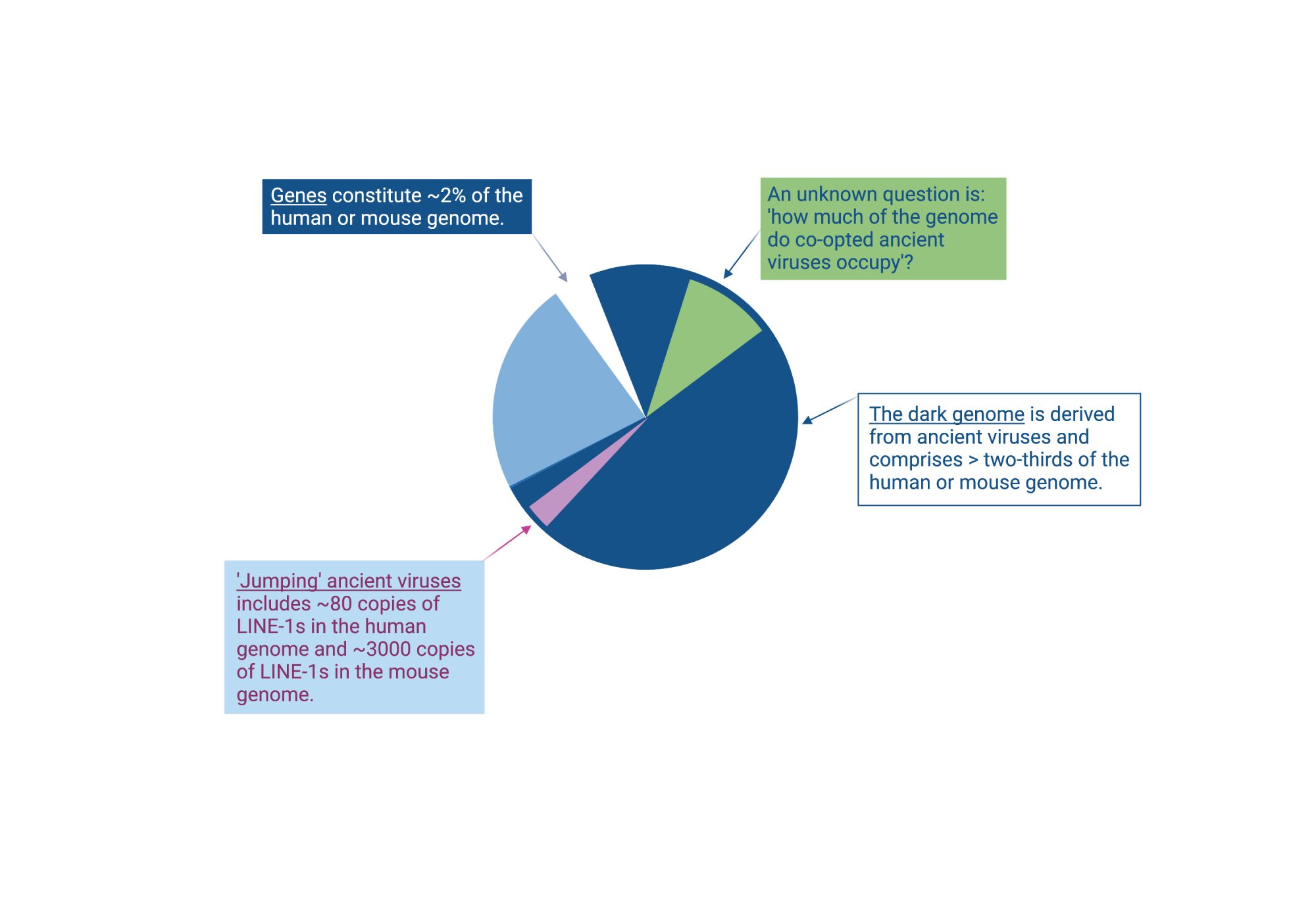
Through a European Research Council starting grant, we have been studying the dark genome in early mouse development. We have aimed to understand which ancient viruses are switched off and on and how and what effect this has on host genes and cell fate. This research has provided new conceptual advances, which will pave the way to research and therapeutic applications in human cells. This article summarizes how ancient viruses that retain jumping activity to this day are regulated and how ancient viruses that are no longer mobile can instead be repurposed for the hosts’ benefit as regulatory elements for our own genes.
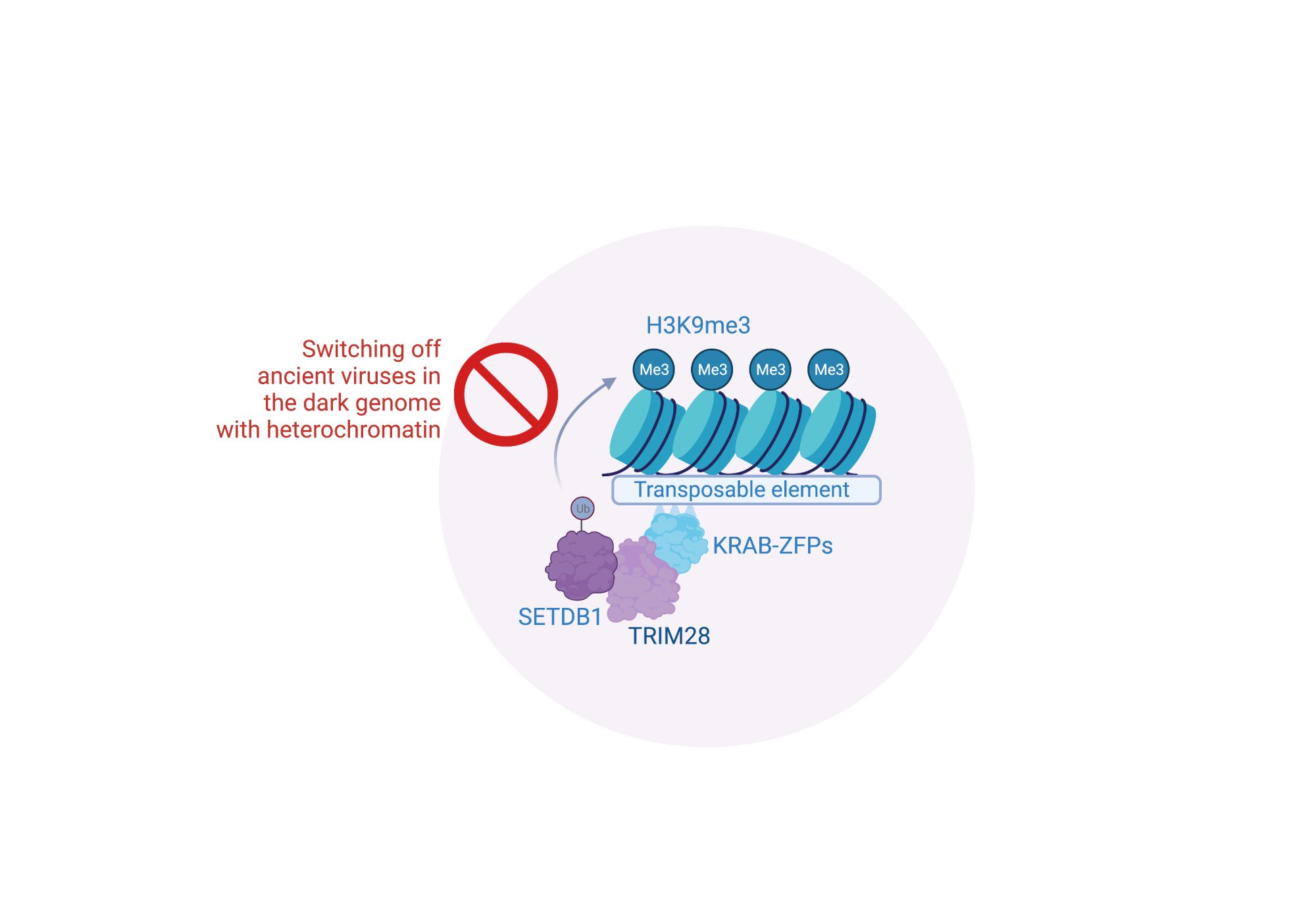
How do we switch off jumping viruses and render them harmless?
In previous work, we uncovered a critical host defence pathway involving TRIM28 and KRAB-zinc finger proteins (KRAB-ZFP) that targets and switches off the transcription of ancient viruses. This defence pathway operates at the chromatin level by recruiting an enzyme known as SETDB1, which is a ‘chromatin writer’ and lays down silent chromatin (heterochromatin) at nucleosomes surrounding ancient viral DNA. SETDB1 does this by adding a modification to histone tails of trimethylation at lysine nine on histone three (H3K9me3), an epigenetic mark associated with gene silencing. In our new studies, we have found that the TRIM28 pathway targets dangerous DNA sequences because they confer ancient viruses the ability to jump. This ensures that the ancient viruses which can jump are shrouded in heterochromatin and rendered harmless.
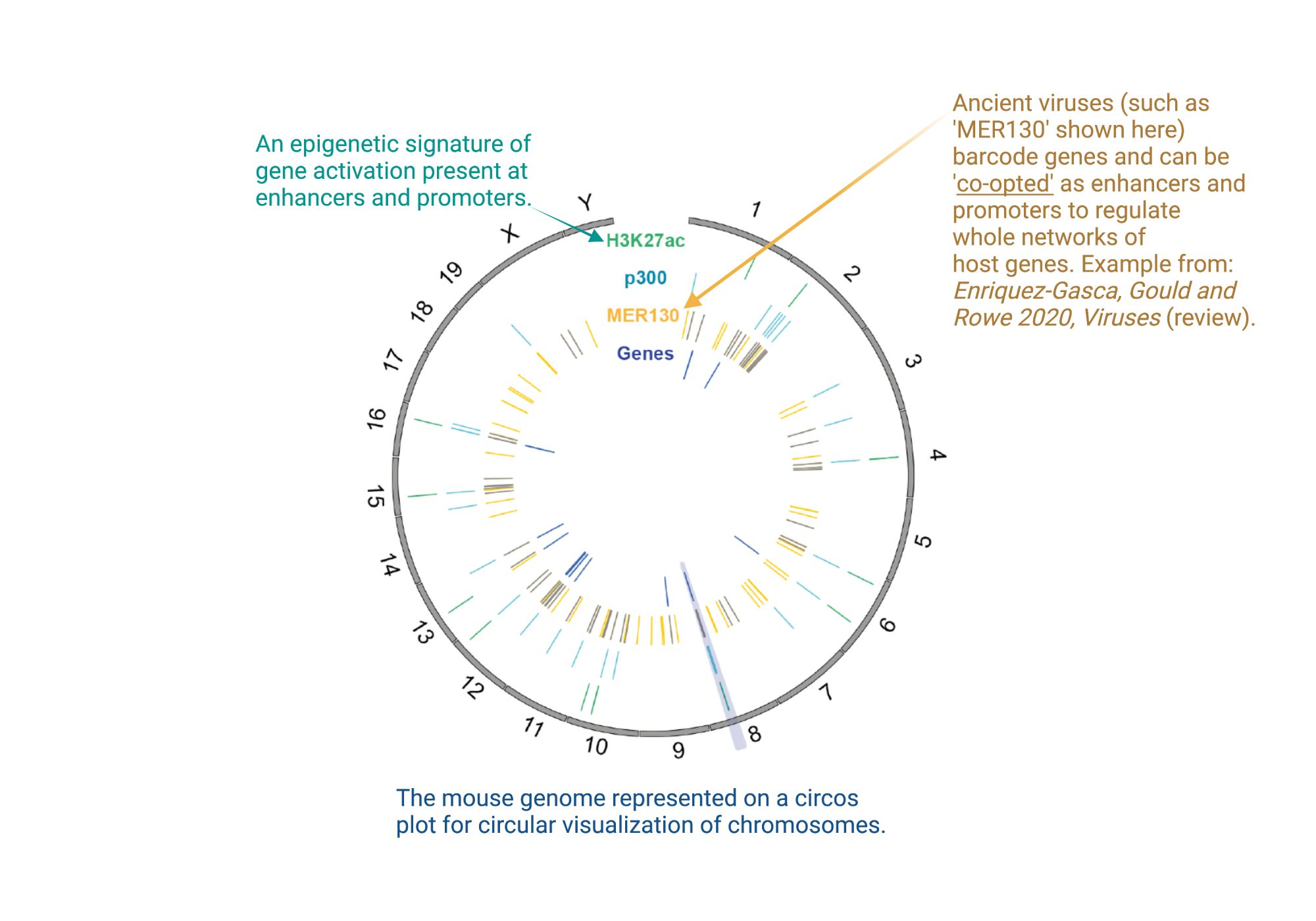
How do we select which ancient viruses to switch on as gene regulatory elements?
Our recent work has also shed light on which copies of ancient viruses in the genome get repurposed by the host to serve as regulatory elements: During the process of retrotransposition, ancient viruses copy or reverse- transcribe their RNA genome into DNA in order to replicate and integrate into new sites within the genome. This process can be error-prone and may lead to mutations and deletions of parts of their sequence along the way. Indeed, our research shows that if they lose their jumping capacity, they also lose vital sequences required for targeting by the TRIM28 pathway. This is because the TRIM28 path targets the very same strings of sequences vital for viral replication (jumping). This indicates that ancient viral copies need to lose their threat to be co-opted by the host as alternative promoters, enhancers and non-coding RNAs.
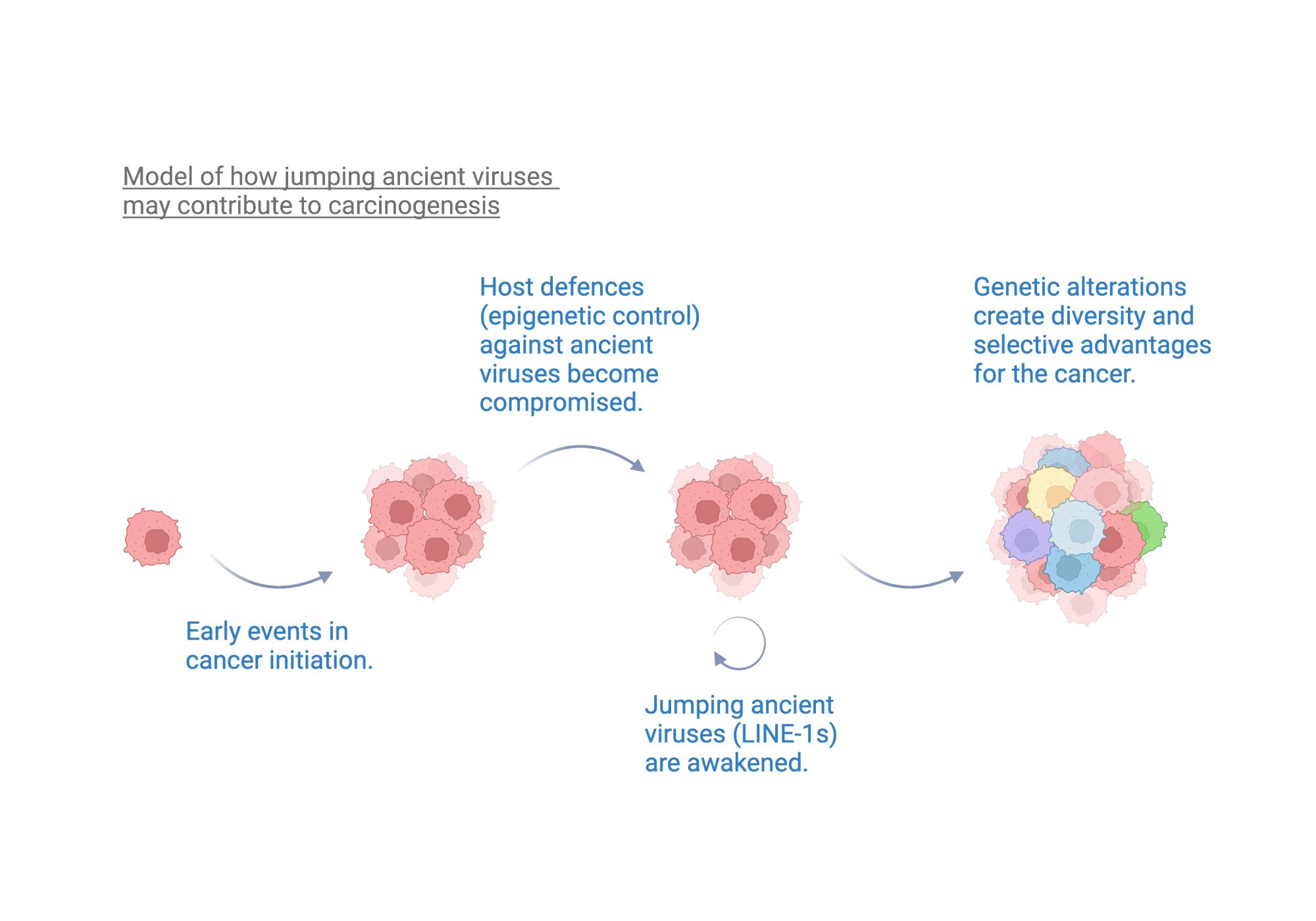
How are these findings relevant to cancer?
As introduced at the beginning of this article, cancer represents a unique disease context wherein multiple layers of host defences are disrupted. This in turn can lead to a loss of epigenetic control of our genomic burden of ancient viruses. This explains why recent research has highlighted an increase in the actual jumping of ancient viruses (from the LINE-1 type) across various human cancers. This jumping activity has been linked to causing diverse genetic alterations in cancers, including mutating tumour suppressor genes (which usually protect us against cancer). We hypothesize that jumping viruses also contribute to cancer evolution by creating increased genetic diversity. This then provides a platform from which groups of cancer cells (clones) can develop with selective advantages, for example, which gain the ability to evade destruction by the immune system. Further research will shed light on new molecular vulnerabilities that can be targeted to outwit cancer.
See https://www.mdpi.com/1999-4915/12/10/1089 for our open access review on this topic.

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.