Here we explore purple non-sulfur bacteria (PNSB) and some of its biotech applications, with a focus on how these applications have been enhanced by manipulating the flow of reducing power
Cellular metabolism refers to the sum of chemical reactions that cells use to obtain energy and material to live, grow, and make products. Many of these reactions are performed by specialized proteins called enzymes, which act on a specific substrate, or input, to produce a specific effect or product, also called a metabolite.
To make metabolites, you need three major inputs: carbon, energy, and electrons. Depending on the organism, carbon can be fixed from atmospheric CO2, or can be obtained from organic compounds like carbohydrates or lipids. Energy can be generated in a variety of ways, but typically results in the formation of phosphate bonds in molecules like adenosine triphosphate (ATP) for widespread use in the cell.
As for electrons, they can cause problems in the cell if they are free or on a highly reactive molecule, so they are often bound to a stable carrier molecule for movement in the cell.
Electrons bound in a usable form are referred to as reducing power, which is generated via electron transfer, or “redox” reactions, wherein the molecule giving the electron(s) is oxidized, and the molecule receiving the electron(s) is reduced. This process is critical for energy management in any cell, as by controlling the movement of electrons (rather than allowing an explosive decomposition of high-energy compounds), the cell can efficiently harvest energy and direct it to a multitude of pathways.
Metabolic pathways are specific series of chemical reactions that cells use to transform one metabolite into another, often either producing or consuming energy. The electron transport chain, for example, uses electron carriers such as nicotinamide adenine dinucleotide (NAD+) to ultimately create a proton gradient for ATP generation. This lets cells convert the energy locked in more energetic chemical bonds (such as those within a molecule of the sugar glucose) into a form that can be more easily used and transported within the cell.
Many other pathways instead result in a reduced product containing more electrons per carbon relative to the substrate, and thus require the input of reducing power. Most “building-up,” or anabolic, reactions that create a complex product necessitate the input of electrons to provide energy or increase the thermodynamic favorability of the reaction. Many metabolic pathways in the cell share enzymes and intermediate products, which results in a highly interconnected set of metabolic pathways that require fine-tuned regulation and direction of limited resources.
By controlling the flow of electrons—and thus reducing power—using redox- balancing pathways, the cell is able to control what, when, and how much of any given product is generated. Cells often do this by controlling the production of enzymes at the genomic level.
In simplified terms, if you consider a genome to be a manual for making all the tools (enzymes) to make products in a cell, then a gene would be the individual instructions for just one of those tools. The cell must write the individual instructions out (transcription) and send them to machinery that can read the instructions and make the tool (translation) before the tool can be used in a pathway to make the product.
Cells can control each aspect of this process through methods such as preventing transcription of a gene to limit product formation, or increasing translation to make more tools. The exciting part about this system is that scientists have also learned how to give instructions to the cells to tell them what (or what not) to do through genetic engineering. This allows us to co-opt the abilities of the cell for human interests, such as the production of goods and services—what we call biotechnology.
The growth of biotech and its value across sectors
Biotech has become increasingly relevant in the modern era as society seeks novel solutions to complex issues like sustainability, advanced medical treatments, and resource management. Overall, there is a growing demand for a more sustainable economy to replace the current petroleum- based model. This is not only due to the devastating environmental and ethical consequences of the current model, but also the increasing global population and decreasing land availability, which highlight the need to come up with sustainable solutions for food production, waste remediation, and energy generation.
By utilizing microbes, which can naturally create and break down their own products, we can close the loop of inputs and products and avoid the dead-end fate of fossil fuel- based products and processes polluting landfills, bodies of water, and the atmosphere.
In the search for microbial species to take on roles as biotech champions, one particular class of microbe shows great promise. Members of the purple non-sulfur bacteria (PNSB) group possess some of the most versatile metabolic capabilities in the microbial world, allowing them to grow under a wide range of conditions—including industrially relevant ones. They have a multitude of native pathways that are suitable for biotech, and several redox-balancing pathways that afford them flexibility in managing cell directives.
Using genetic engineering, scientists can alter these directives to enhance the production of our interests- for example, biotech applications.
Purple non-sulfur bacteria and its applications
In this article, we provide a brief overview of PNSB and some of their biotech applications, with a focus on how these applications have been enhanced by manipulating the flow of reducing power.
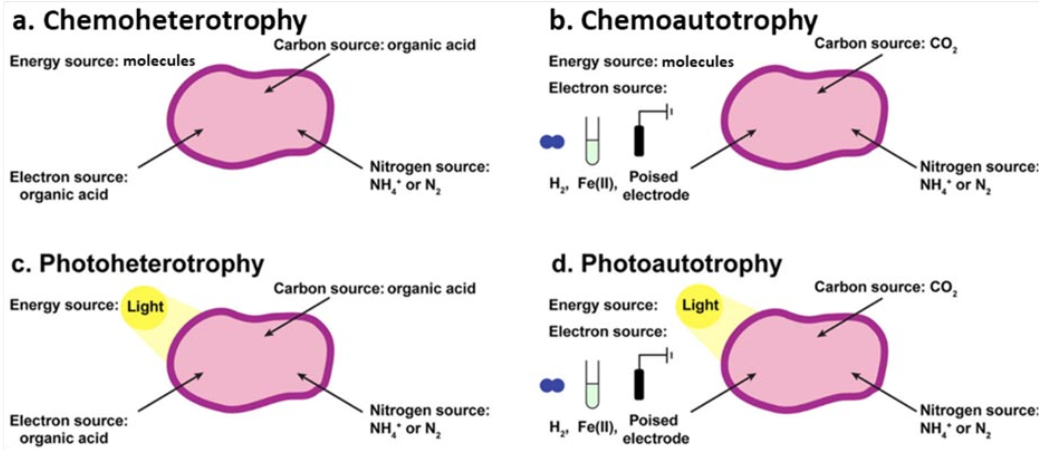
Purple non-sulfur bacteria (PNSB) are a group of gram-negative bacteria with an incredibly diverse set of metabolic capabilities. They can grow using each of the four primary metabolic modes (photoauto-,photohetero-, chemoauto- and chemoheterotrophy), meaning they can obtain energy from light (photo) or chemical compounds (chemo), and carbon from organic (hetero) or inorganic (auto, CO2) sources.
They can grow in the presence or absence of oxygen (aerobic and anaerobic, respectively), and can even fix their own nitrogen (N2 to NH3) in addition to performing specialized processes such as aromatic compound degradation.
Altogether, this metabolic versatility affords purple non-sulfur bacteria a great degree of flexibility in adapting to variable nutrient and environmental conditions, thus outcompeting less versatile microbes. One of the more interesting traits of PNSB is that some species can obtain electrons via extracellular electron uptake (EEU).
Extracellular electron uptake
For EEU, they can oxidize metals such as soluble iron(II) to insoluble iron(III) to obtain electrons, or even directly uptake electrons from a cathode. This has been
best characterized in model species Rhodopseudomonas palustris, where the Bose Lab identified the pio (phototrophic iron oxidation) genes encode an electron conduit in EEU. Under photoautotrophic conditions, they observed that EEU relied on the photosynthetic electron transport chain (pETC) to convert electrons into usable reducing power. Further, the pETC is linked to the fixation of CO2 via the Calvin- Benson-Bassham Cycle (CBB), where the latter consumes reducing power generated by the former. Thus, the cell can power both energy generation and carbon fixation.
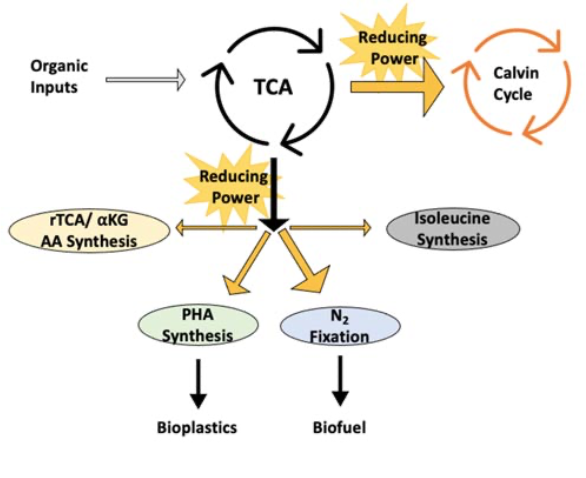
PNSB’s metabolic flexibility is in part, thanks to their variety of ways to direct reducing power, referred to as redox-balancing pathways. It is critical for the cell to maintain control of the ratio of reduced to oxidized cofactors (ie., NADH/NAD+), as some reactions require the thermodynamic favorability generated by a high ratio to drive the reaction forward. However, too much reducing power can generate a toxic environment in the cell. Thus, the cell requires pathways that will consume this excess reducing power, as might be generated during growth on a more reduced substrate relative to biomass.
The major strategy utilized by PNSB is the Calvin-Benson-Bassham Cycle (CBB), which fixes CO2 into a usable carbon intermediate for central metabolism, like glycolysis or other carbon management pathways. CBB typically handles 40-60% of redox flux in PNSB, as the cell favors the carbon efficiency of using excess reducing power to fulfill a cell requirement, and CBB is usually the most thermodynamically favorable option.
Nitrogen fixation
Another available pathway is nitrogen fixation, or specifically the nitrogenase enzyme, which consumes electrons and protons (H+) in order to fix atmospheric nitrogen to bio-available ammonia, producing hydrogen gas as a byproduct. This enzyme can also function solely as a hydrogenase in the absence of a need for fixed nitrogen, allowing it to continue acting as a means to consume reducing power. However, nitrogenase is extremely energetically expensive to run (as well as requiring anoxic conditions), and is thus favored less compared to CBB.
A third option is the production of polyhydroxyalkanoates (PHA), which are polyester bioplastics produced by the cell to store carbon under nitrogen-limited conditions. These polyesters can then be broken down when the cell needs carbon, much like humans might store and consume fat for future energy needs. Again, however, the limitations on what conditions allow for PHA production usually mean this is a less favored means of consuming reducing power.
Alternative pathways to consume reducing power
In addition to these broad-spectrum purple non-sulfur bacteria pathways, some species are also observed to use specific alternative pathways to consume reducing power. This trait has been primarily characterized in Rhodospirillum rubrum and hinted at in other species, but further research is required to confirm its prevalence. Species with these alternate pathways can utilize a specialized enzyme to run part of the tricarboxylic acid cycle (TCA) in reverse to produce alpha-ketoglutarate as a precursor to several amino acids. Similarly, the synthesis of the amino acid isoleucine was also implicated in redox balancing. However, because the consumption of reducing power is limited by the demand for the amino acids produced by these pathways, they are unlikely to play a major role in redox balancing when other pathways are available.
PNSB possess several biotechnologically relevant native pathways, most of which are directly implicated in redox balancing. Thus, several groups have had success in increasing yield or activity by influencing the accompanying redox pathways, either by knockout, overexpression, or introduction of a pathway from an entirely new source. One such application is the production of biofuels, which simply refers to fuel derived from biomass. PNSB have been demonstrated to be capable of producing several types of fuel, with the most popular product arguably being biohydrogen.
H2 is clean-burning, energy-dense, and when produced by PNSB, quite pure compared to traditional purely chemical or physical methods that contaminate the product with other gases. In addition to being produced as a byproduct of nitrogen fixation, it can also be generated by several dedicated hydrogenases, which only produce H2. However, although PNSB is highly efficient at producing H2, since nitrogenase is energy-expensive to run the cell will typically limit activity in favor of CO2 fixation.
To get around this, several groups have taken the approach of removing alternate electron-consuming pathways to force the cells to divert the excess redox potential into hydrogen production. Wang et al. saw that the elimination of the gene for RuBisCO, the primary CO2 fixing enzyme, increased the production of hydrogen in R. rubrum. Yang & Lee knocked out phbC, encoding a PHA-producing enzyme, in R. palustris and saw a 4% increase in H2 production when grown on acetic acid. Taking an alternate approach, Ma et al. sought to over-express fdxN in Rhodobacter sphaeroides, which encodes ferredoxin I: the main electron carrier donating to nitrogenase. They found that by increasing the pool of available electron carriers, they could increase the rate of hydrogen production.
As for other biofuels, the Bose Lab has recently demonstrated the ability of R. palustris TIE-1 to generate n-butanol under autotrophic growth, which was further enhanced by the deletion of genes involved in the nitrogen-fixing pathway.
This last finding employs the critical concept of bio-engineering: that reducing power is a universal substrate, and that when you create an available pool by knocking out other pathways that consume reducing power and introduce a new pathway to consume it, the cell is likely to utilize it. This concept is what allows scientists to swap in pathways that may not be native to that species, as was done with n-butanol.

Another biotech application is the production of PHAs, specifically polyhydroxybutyrate (PHB), which is currently being studied as a bioplastic: a sustainable replacement for petroleum-based plastics. Although they currently lack the thermal stability and lower crystallinity of modern petrol-derived plastics, they are being continually optimized through methods such as improving co-polymerization, which increases flexibility. As with biofuels, given that PHA production is used by the cells for redox-balancing, manipulating the electron flow is one way to increase production.
However, it serves the reader well to remember that these redox pathways are functioning to do more than consume reducing power, and their original function can have overriding consequences. As an example, the Bose Lab found that by overexpressing RuBisCO in R. palustris TIE- 1, the cell would increase production of PHB due to the increased flow of fixed carbon, despite the over-active Calvin cycle consuming more reducing power. Similarly, multiple groups saw that using different carbon sources or a combination could improve PHB production depending on the species.
Challenges and opportunities for biotechnological applications
The metabolic flexibility of PNSB presents as many challenges as it does opportunities when attempting to utilize the bacteria for biotechnological applications. PNSB are quite skilled at adapting to changing conditions, whether that be the loss of a nutrient or the loss of a pathway, and they frequently respond in unexpected and exciting ways.
On one hand, this provides us with more information on the global metabolism and how PNSB might respond under certain growth conditions. On the other hand, this makes it difficult to artificially control the flow of reducing power, as scientists must account for many more variables and pathways that PNSB may engage.
For this reason, several groups have developed metabolic models using a combination of experimental data and powerful computational software to better predict purple non-sulfur bacteria behavior and identify carbon and electron flow. These models are constantly improving and expanding with newfound observations, and will undoubtedly be critical for the optimization of PNSB biotechnological applications.
Here in the Bose Lab, we continue to define the bio-engineering capacity of PNSB by investigating how electron flow can be utilized to enhance production and growth for biotech applications. The reader is encouraged to see other Ebooks in this series for details on how purple non-sulfur bacteria and other electroactive microbes can directly uptake electrons for product synthesis.
We thank Brian Gallagher for editing this eBook.
Read and download the full version here.

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.


