In this article, Antoinette van Ouwerkerk and Salvatore Spicuglia from INSERM highlight the significance of regulatory variants within the non-coding genome – often referred to as the ‘dark genome’ – in influencing gene expression and disease
The major genetic cause of human disease has historically been thought to be mutations within coding genes. However, there has been a shift in perspective over recent years, with more attention now being paid to the role of the non-coding regions in the genome. Intense research in this area has revealed that the non-coding genome contributes to gene-regulatory networks that serve to control where and when sets of genes are expressed. In turn, dysregulation of gene expression due to changes or alterations in the non-coding genome drastically impacts the aetiology of different diseases.
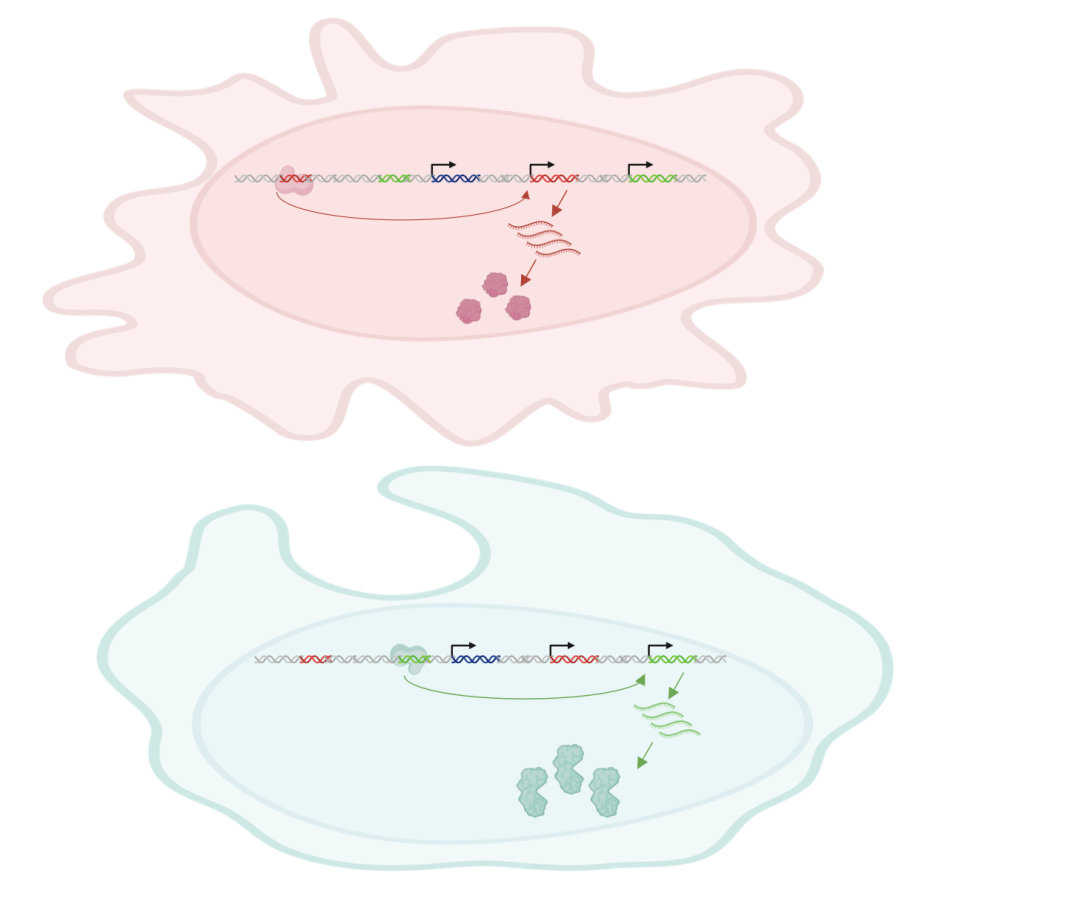
The dark genome: cis-regulatory elements and the control of gene expression In the vast landscape of our genetic makeup, there is a mysterious territory often referred to as the ‘dark genome’. This territory does not code for proteins like the more familiar parts of our DNA but holds the information on how our genes are expressed. At the heart of these enigmatic domains lie cis-regulatory elements, molecular switches that act like conductors orchestrating the symphony of gene expression. Unlike the genes themselves, which provide the blueprint for proteins, these elements control when and where genes are turned on or off. For example, a cell that functions in the immune system, such as B-cell lymphocytes, will express a different subset of genes than a hepatocyte in the liver.
Cell-type-specific regulation of gene expression is achieved through the interaction between regulatory elements that are either proximal (called promoters) or distal (called enhancers) to genes. They are short stretches of DNA that contain binding sites for a type of protein called transcription factors, which can increase the expression of nearby target genes. Enhancers, which primarily function in a cell type-specific manner, play an important role in determining where and when genes are expressed in different cells in the body (Figure 1). Understanding the intricate dance of these cis-regulatory elements sheds light on the complex mechanisms governing our biology.
Genetic variation within the non-coding genome plays an important role in disease
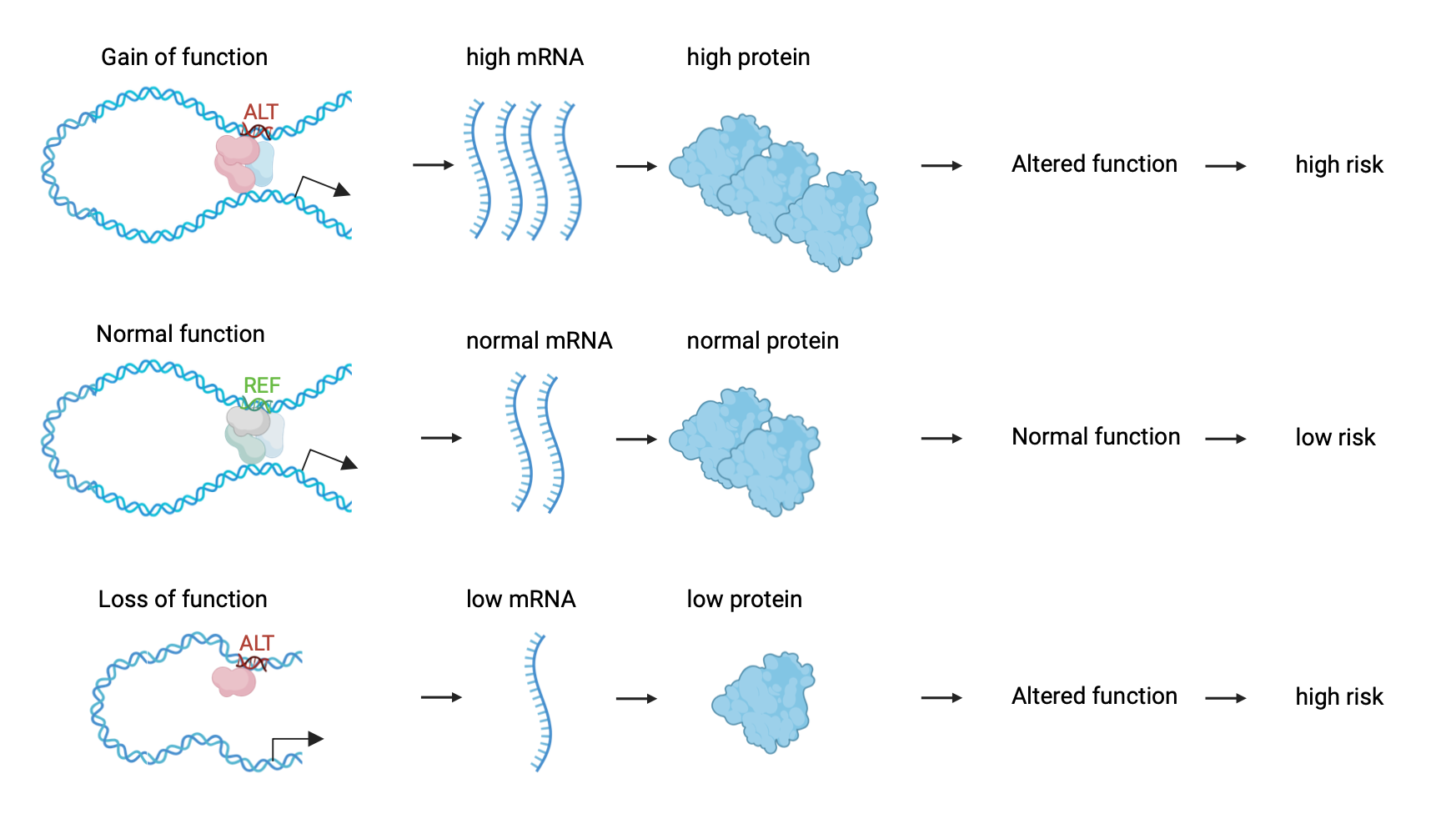
While the coding regions of our DNA garner much attention, it’s becoming increasingly clear that genetic variations in the non-coding genome, or the dark genome, can disrupt the finely tuned control of gene expression, leading to disease susceptibility or progression. Cis-regulatory elements, and in particular enhancers, can be altered in different ways, such as through structural variation or point mutations, which lead to dysregulation in gene expression and, therefore, increase the susceptibility of the carrier individuals to a particular condition or disease (Figure 2). With this in mind, the EU-funded ENHPATHY project was built to investigate different topics around the role of enhancers in health and disease. (1) By shining a spotlight on this previously overlooked landscape, researchers are uncovering a treasure trove of information that promises to illuminate the intricate molecular mechanisms behind a myriad of diseases, offering new avenues for diagnosis, treatment, and prevention.
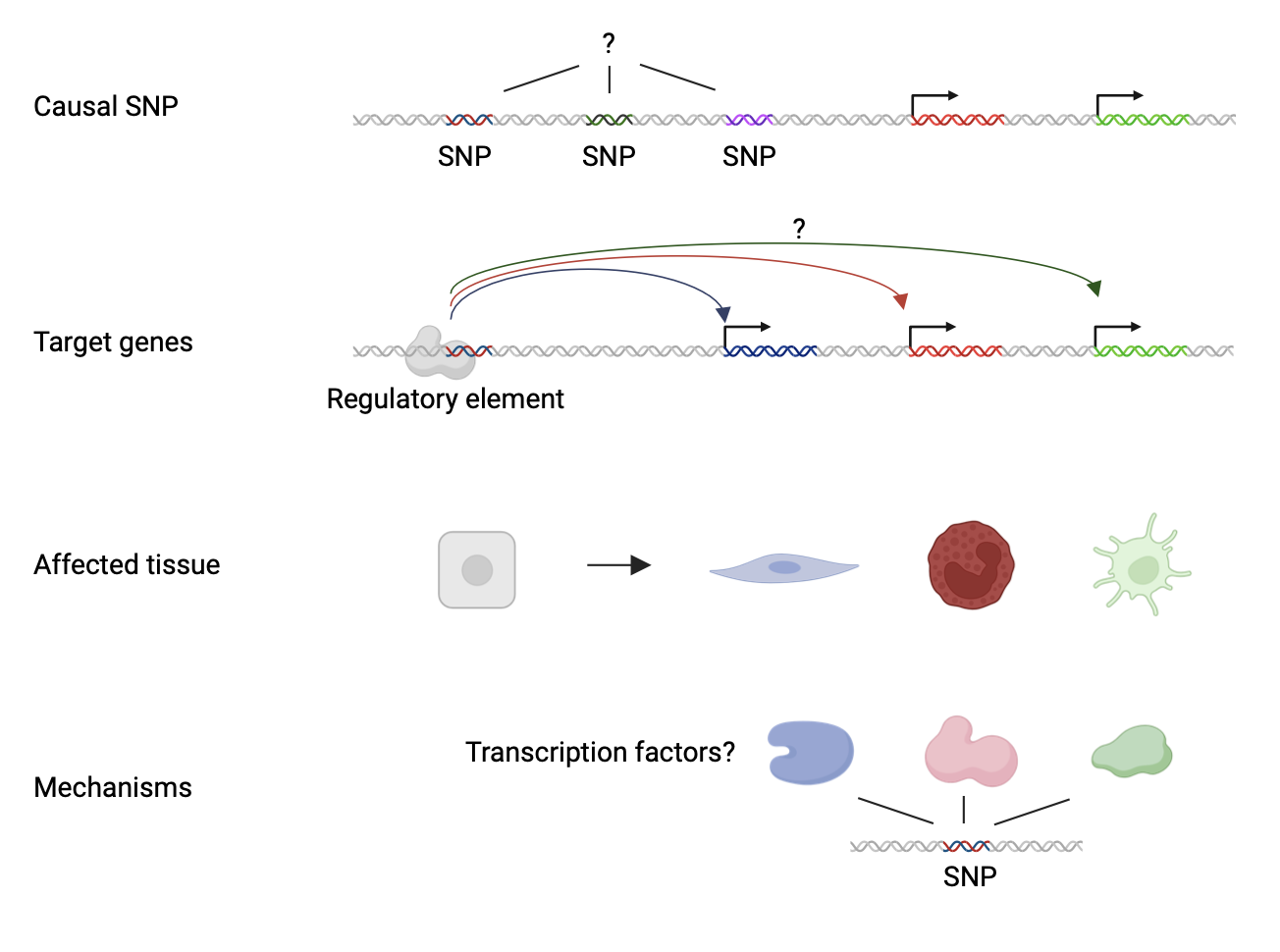
Challenges in understanding the role of regulatory variants
Delving into the world of regulatory variants within the dark genome presents a formidable set of challenges for researchers. This is a highly complex area of research, as identifying and characterizing human variation in DNA regulatory sequences is a technically challenging task (Figure 3). Unlike mutations in coding regions that directly alter protein structure, regulatory variants often exert their effects in more subtle ways, making them harder to detect and interpret. Additionally, the sheer complexity of gene regulation means that pinpointing the precise functional consequences of these variants can be akin to navigating a labyrinth in the dark. Compounding these challenges are the vast amounts of data generated by high-throughput sequencing technologies, requiring sophisticated computational methods to go through the noise and identify meaningful signals. Despite these obstacles, the allure of unravelling the mysteries of the dark genome and its regulatory variants continues to drive research forward, holding the promise of transformative insights into human health and disease. In this regard, the ENHPATHY consortium has recently provided an insightful perspective to pinpoint the critical areas of investigation to further our understanding of cis-regulatory dysfunction in disease. (2)
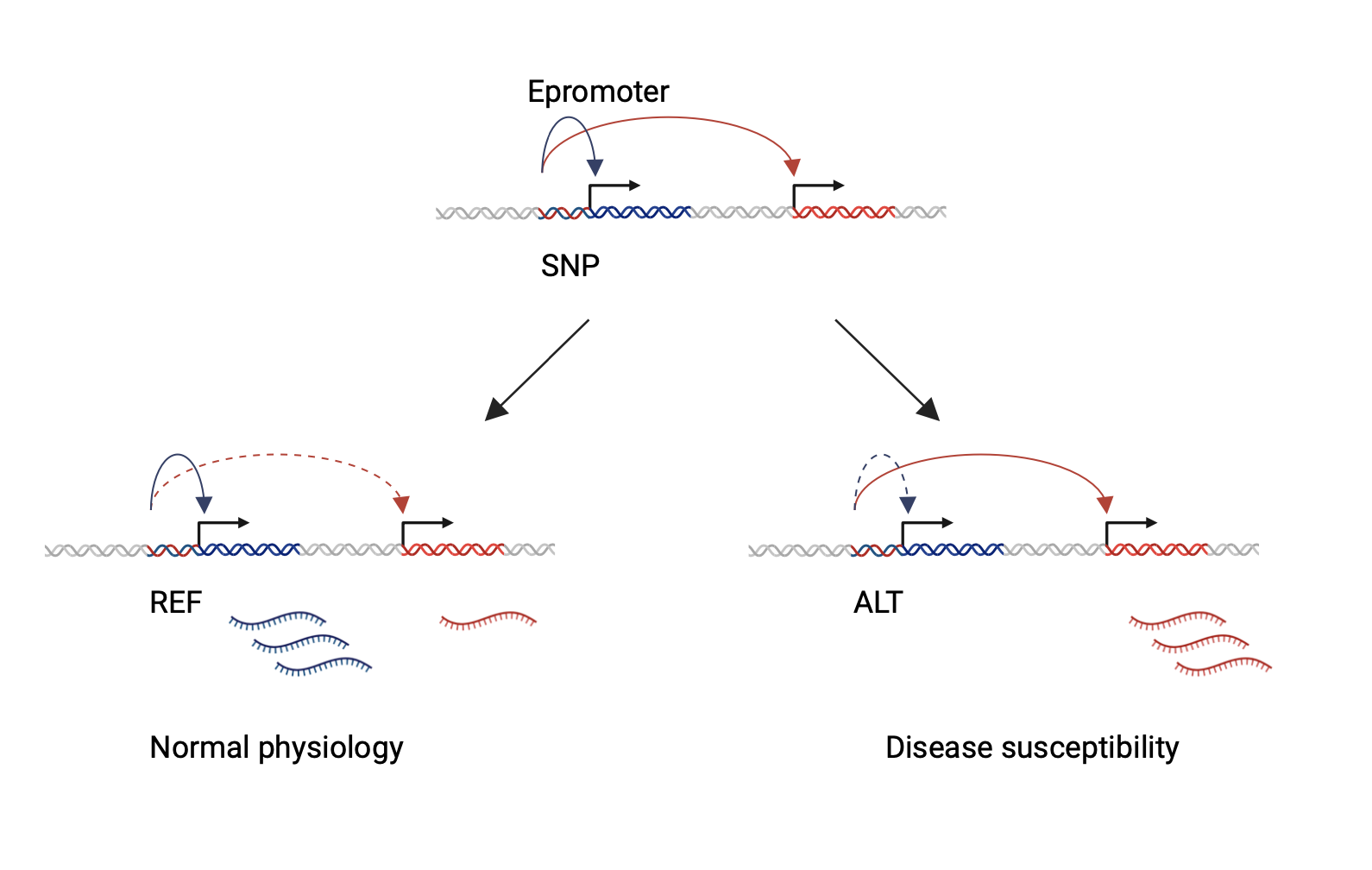
The confounding role of Epromoters in disease
Recent studies have reported that a subset of promoters, termed Epromoters, also work as enhancers to regulate distal genes. This new paradigm opened novel questions regarding the complexity of our genome and raises the possibility that genetic variation within Epromoters has pleiotropic effects on various physiological and pathological traits by differentially impacting multiple proximal and distal genes (Figure 4). Thus, Epromoters might play an important role in the regulatory landscape and represent major contributors to phenotypic variation and disease. (3) Current work in our laboratory aims to elucidate the molecular mechanisms of these puzzling regulatory elements.
Acknowledgements
Preparation of this article was done with the support of the ‘Enhpathy’ Marie Sklodowska – Curie actions (MSCA) – Innovative Training Network (ITN) funded by the European Union’s Horizon 2020 research and innovation program (Grant Number: 860002), an MSCA postdoctoral fellowship (Eprom-101065610) and the ANR-funded EnhProm project (ANR-23CE12-0008-01).
References
- Spicuglia, S., and Rada-Iglesias, A. (2023). Understanding enhancer function to understand human disease. Bioessays 45, e2300149. https://doi.org/10.1002/bies.202300149
- Zaugg, J.B., Sahlén, P., Andersson, R., Alberich-Jorda, M., de Laat, W., Deplancke, B., Ferrer, J., Mandrup, S., Natoli, G., Plewczynski, D., et al. (2022). Current challenges in understanding the role of enhancers in disease. Nature Structural & Molecular Biology.
- Malfait, J., Wan, J., and Spicuglia, S. (2023). Epromoters are new players in the regulatory landscape with potential pleiotropic roles. Bioessays 45, e2300012. https://doi.org/10.1002/bies.202300012.

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.