Dr Joanna Leng, School of Computing, University of Leeds, UK, presents a project from the fellowship, on the PERPL (pattern extraction from relative positions of localisations) software which analyses super-resolution light microscopy (SRLM) data
In 2018, I started a Research Software Engineering Fellowship. RSE is an emerging role. Here I present a project from the fellowship on the PERPL (pattern extraction from relative positions of localisations) software which analyses super-resolution light microscopy (SRLM) data.
RSE and the Innovation Pipeline
The e-Infrastructure theme of EPSRC, the Engineering and Physical Sciences Research Council, which provides world-leading research equipment and facilities for the natural sciences, funded this fellowship. Around 2010 they realised a critical issue: a need for better quality software in the UK academic research environment.
A campaign to raise awareness of this was started, and the term ‘RSE’ was created. By mid-2010, they realised that RSEs were under-represented in senior roles and academic duties, e.g., the peer review of funding applications. The RSE Fellowship scheme was developed partially to correct this and get RSEs into the academic decision-making processes.
The full diversity of the RSE role has yet to be fully understood; however, e-Infrastructure links with research facilities, including HPC (High-Performance Computing) services with dedicated software developers. Therefore, the RSE campaign engaged more with RSEs that were more likely to optimise software for HPC, but RSE working on early innovation software (and not for HPC) is also needed. My experience places me closer to the start of the innovation pipeline, so my fellowship is based on early innovation software (Figure 1).
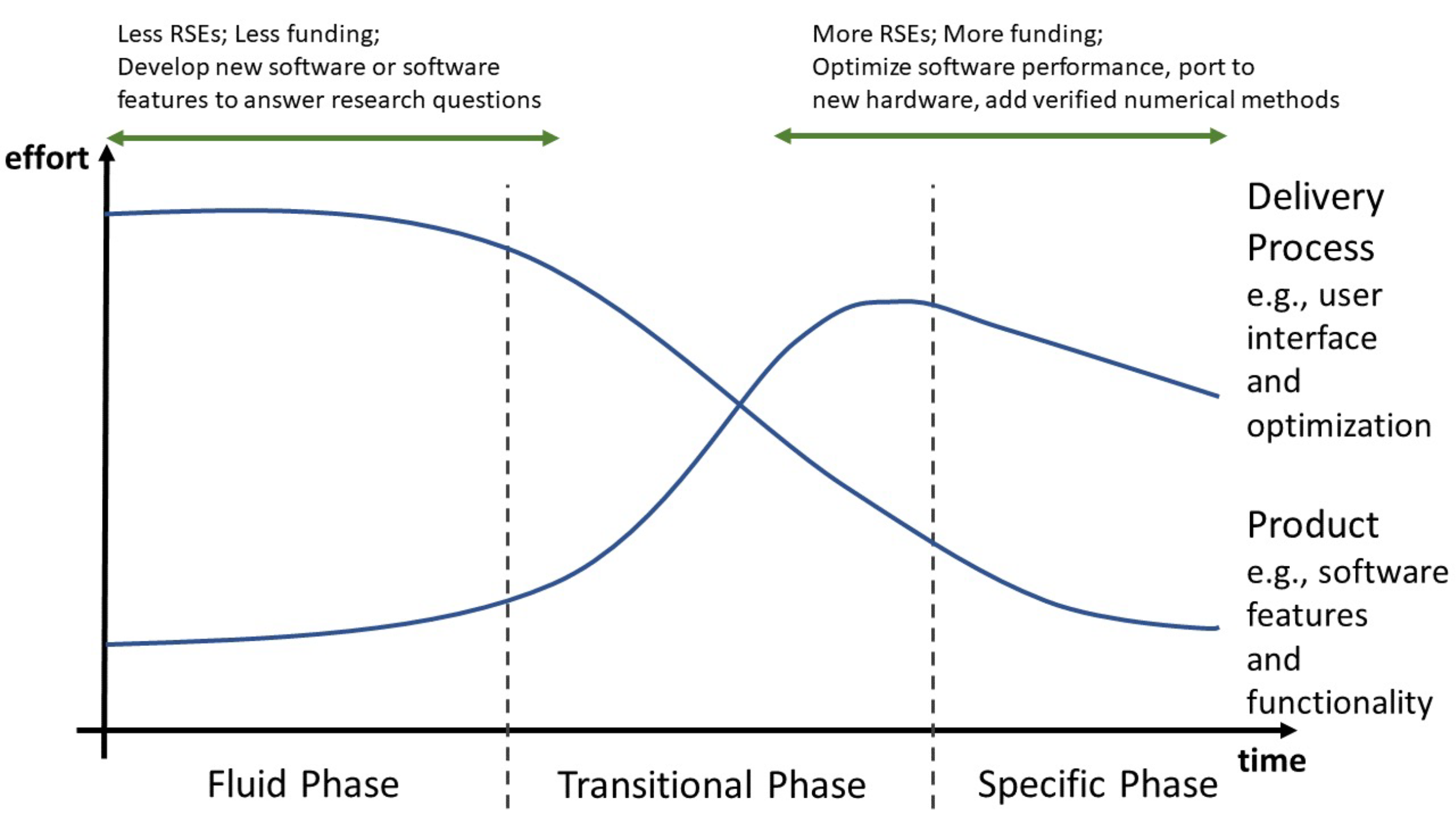
1) the Fluid Phase, where flexibility is needed because of uncertainty in the product idea, the technology and the market and, this case, the needs of the research community; 2,) Transitional Phase when the technology, the application and the customer’s needs are better understood until a ‘dominant design’ emerges; 3,) the Specific Phase the ‘dominant design’ shifts from being different to having good performance
SLRM and Software Innovation
Dr Alistair Curd is a physicist who builds SRLM that surpasses the conventional limits of light microscopes. As a result, SRLM is potentially a disruptive technology in medical assessment. However, software to analyse these molecular distributions is limited.
Therefore, Alistair initially developed PERPL on his own to increase the application and impact of SRLM. PERPL assesses molecular distributions for regular patterns to improve the biological understanding of a specimen.
Collaboration on PERPL
Alistair developed the initial capabilities of PERPL with knowledge of the Python programming language. Still, with limited experience in software engineering, he realised it was becoming difficult to extend and for others to use. As the software and the underlying science were complex, the best way forward was for Alistair and me to work together. However, this close collaboration involving teaching software engineering skills is not a commonly recognised RSE practice.
Easy-To-Extend & Easy-to-Use
We improved the source code in ways generally accepted by software engineers and RSEs to make it easier to re-use, which also makes it easier to share and extend. This included moving the source code to a version-tracking service (BitBucket), adding software tests and automating documentation.
The source code was not modular, and the naming conventions needed to be more explicit; improving these were vital to making it easy to extend. We used automated analysis to implement a Python programming standard style (PEP8), making it easier to understand, share and grow.
We drew semantic maps of the microscopy technique, the data, the aims of the analysis, the results and the importance of each part of the results. It was slow, but necessary because SRLM and PERPL were in the Fluid Phase (Figure 1), and the terminology needed to be more meaningful. Once we had good conceptual models, we agreed on the data structures and naming conventions that resulted in easy-to-read code. This type of software re-engineering is expected early in the innovation pipeline when there is much uncertainty about the product. However, it is not a funded or generally recognised RSE practice.
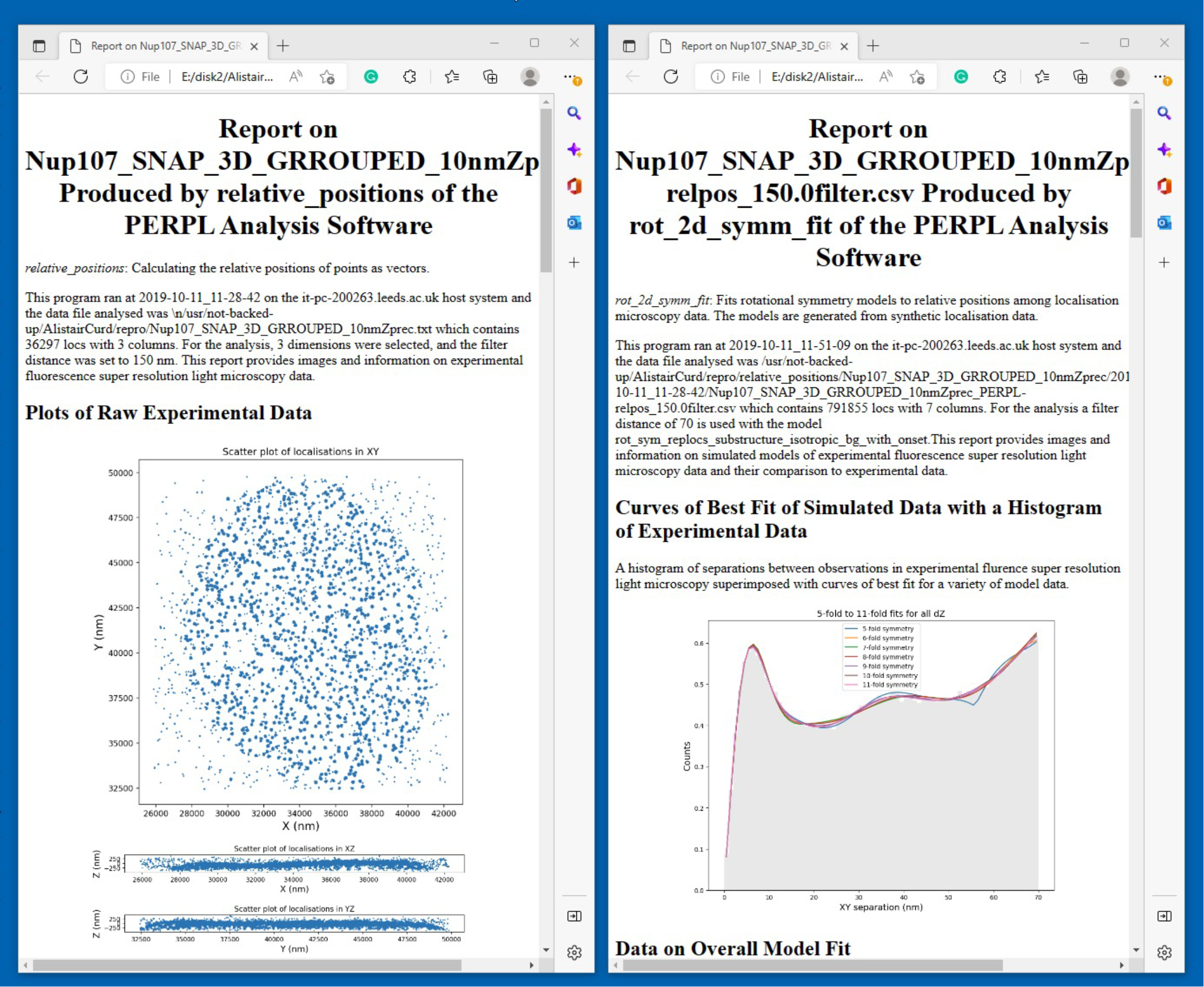
We made PERPL easy to install and simplified the command line user interface. We introduced automatic HTML reports to present results with graphs, pictures, tables and text, necessary for its scientific use. Finally, we shared the software with Prof Michelle Peckham, a cell biologist and expert in SRLM, who beta tested it. Alistair selected data that is available on BitBucket. After a year of collaboration, the results of PERPL were ready for publishing https://doi.org/10.1021/acs.nanolett.0c03332.
The Future of PERPL
After this publication, Alistair wanted to become a RSE and now has a 1-year fellowship with the Wellcome Trust, developing new analyses for SRLM data. In late 2021, Prof Philip Quirke, a senior medical researcher in cancer and a medical AI research student, Oliver Umney, joined the team. Oliver has embedded PERPL analysis in deep learning to identify the patterns within SRLM data on cancer treatments.
In 2020, Alistair began a collaboration using PERPL to assess the quality of a major new SRLM technique https://doi.org/10.1038/s41592-022-01694-x, with Kirti Prakash (Institute for Cancer Research, ICR). They plan to use it to guide SRLM experimental protocols. Emre Kose, a pathologist and analytical scientist from ICR, is developing a prototype of PERPL with a graphical user interface in Streamlit, which quickly generates ML/AI web-apps.
PERPL has two steps, analysis and modelling, which require human interaction between them. The aim is to improve the modelling and introduce automation between the steps. This will improve its functionality making it more “dominant” and moving it along the innovation pipeline.
Software is relatively new to the research environment, and new software is essential to boost research outputs, develop new technologies and materials, and reach societal goals. We urgently need to think how to improve its development and adoption. This project demonstrates one-way early innovation software can be developed.
References are available on request.
Acknowledgement
Thanks to Dr Alistair Curd, Prof Michelle Peckham, School of Molecular and Cellular Biology, University of Leeds. This work was supported by the EPSRC grant EP/R025819/1, Wellcome Trust grant 204825/Z/16/Z and BBSRC grant BB/S015787/1.

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.


