Emmanuel Kifaro from the SACIDS Foundation for One Health, and College of Veterinary Medicine and Biomedical Sciences, Sokoine University of Agriculture, Tanzania, details the potential application of microparticles for active surveillance of viral infections from non-invasive animal matrices
The tremendous increase in the occurrence of infectious diseases and drug resistance poses higher risks to global health, but microparticles could aid the surveillance of these pathogens before they spread.
Infectious diseases of viral origin have emerged as potentially dangerous pathogens globally, normally associated with a high death toll in human and animal populations. These pathogens have shown characteristic speed in their evolutionary processes, enabling rapid host immune evasion and maximizing their propensity for fast spread. Some of these pathogens have shown a high ability for cross-species transmission affecting humans and animals of food security importance, with severe consequences on food security and economic collapse of the individual households and the affected regions (e.g Rift Valley fever virus).
Taking the example of the recent SARS-CoV-2, which spread very fast, killing millions of people globally. It showed the huge gap along the diagnostics chain as it presented a severe shortage of testing kits. In such a scenario, sampling and surveillance activities are high risks activities, predisposing health workers to the exposure of lethal infectious agents especially when invasive procedures are mandatory for diagnostics purposes.
Limiting the spread of highly infectious agents at a point source
Looking for another type of matrices for diagnostics is of high importance to limit the spread of highly infectious agents at a point source. Non-invasive matrices (saliva and stool samples) are good candidates to be considered. On the other hand, there is a continuous need for innovation of more situation-based and reliable DNA-based diagnostics test kits.
In the current study, the application of hydrogel microparticles was optimized for direct capture and amplification of viral RNA from saliva and stool samples without prior purification of the nucleic acids.
Non-invasive matrices for diagnostics purposes
Non-invasive matrices are normally obtained through painless procedures without the need to restrain the animal. This sampling method carries several other key advantages over the invasive sampling technique, which is currently the preferred method for the collection of whole blood, swabs, and tissue samples.
The non-invasive matrices are freely available and easily accessed, cheap and simple collection methods, less need for experts for sample collection, and safer sampling method for health care workers and researchers. Despite their complex nature, which includes high levels of PCR inhibitors and debris (e.g. stool). Recently, they have been reported for the detection of several pigs, goats, and birds’ viral pathogens using aqueous phase PCR following nucleic acid purification using commercialized kits. There is evidence for the detection of porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2), from oral fluid (rope saliva) (Prickett et al., 2008), peste des petits ruminants from stool samples of sheep and goat (Bataille et al., 2019), and avian influenza from bird’s droppings (Dhumpa et al., 2011). More recently, SARS-CoV-2 was directly detected by RT-qPCR from saliva samples using thermal heat for viral lysis in place of commercial nucleic acid extractions kits.
Depending on the type of viral pathogen and its pathogenesis. The major challenges facing the use of non- invasive matrices for molecular diagnostics include low template concentration, overcoming the PCR inhibitors, and rapid degradation of the viral nucleic acids and the matrix.
What are the microparticles?
The emergence of nanotechnology come up with numerous types of both nano- and microparticles fabricated from different source materials, including glass, ceramics, and polymers (e.g. hydrogels). They are widely used for various applications in the agriculture, food industry, and health sectors. Hydrogel microparticles are commonly used polymeric materials in the health sector fabricated from either natural or synthetic raw materials such as starch. They provide a wide range of applications from wound healing, drug delivery, and diagnostics due to their unique properties, including porosity, high water content, biocompatibility, and flexibility.
The microparticles for qPCR
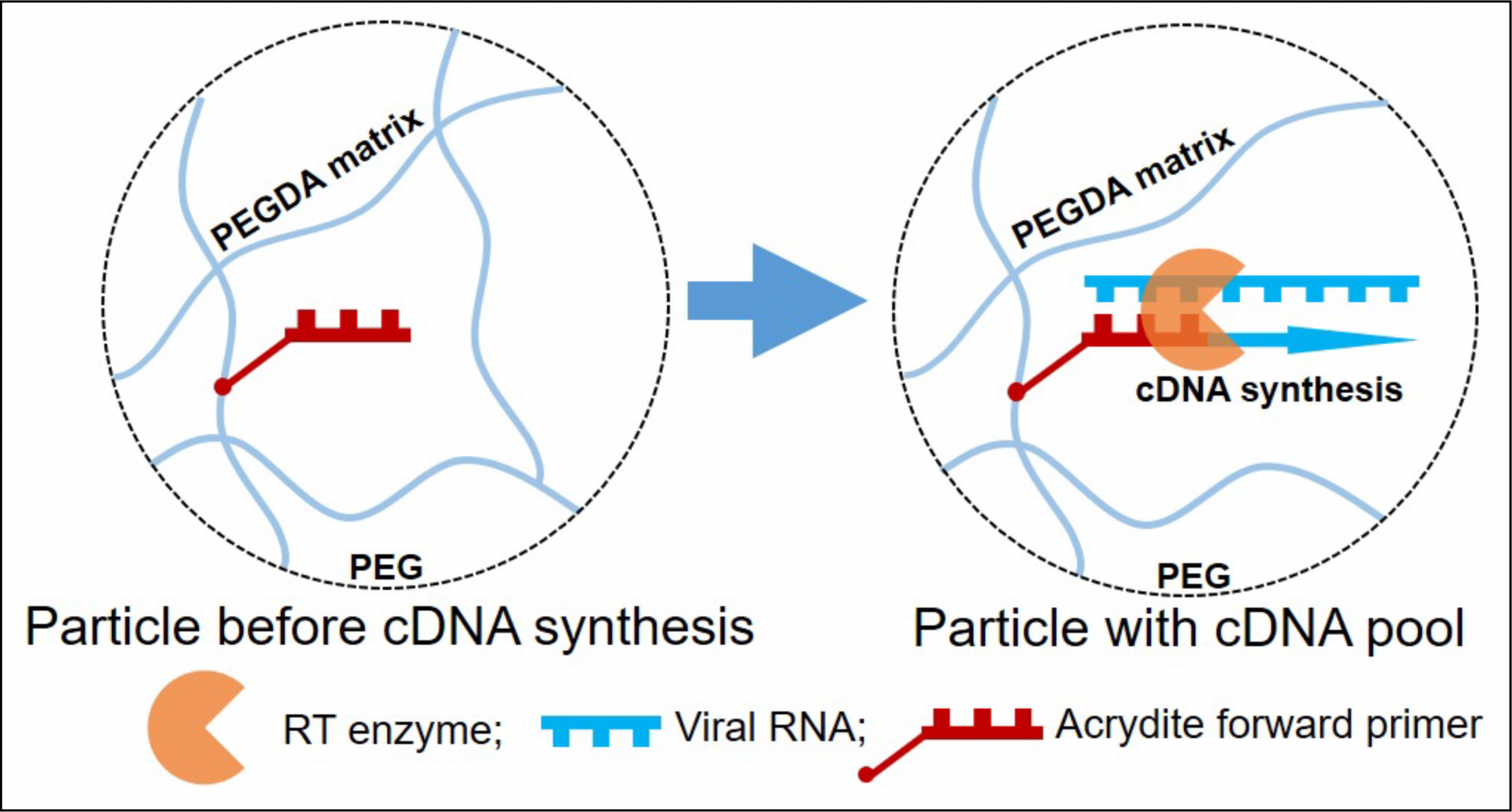
Functionalization of these hydrogel microparticles with various functional groups is the key feature of hydrogel polymers, providing the particles with the versatility required for diagnostics applications.
In this case, the particles were fabricated from poly (ethylene glycol) (MW 600) to provide porosity of particles, and UV-photocrosslinkable poly (ethylene glycol) diacrylate (MW700). The later, facilitate the functionalization of the particles with an acrydite modified forward primer, which is gene-specific and used during the reverse transcription step.
This novel protocol contains only three main hands-on steps, which are viral lysis, cDNA pool synthesis, and qPCR. The protocol begins with heat treatment of the matrices for viral lysis and inactivation of heat-sensitive PCR inhibitors from the matrices at 100°C for up to 10 minutes. Then, at 42°C for 10 minutes, the covalently immobilized primer facilitated the direct capture by hybridization of viral RNA, and direct synthesis of complementary DNA (cDNA) forming a cDNA pool inside microparticles.
The formed cDNA pools inside the particles can be used immediately as a source of template for qPCR or can be stored at room temperature for more than two months for later use. The technique was capable of direct detection of influenza A virus and infectious bronchitis virus spiked into saliva and stool samples using hydrogel microparticles without prior purification of viral nucleic acids from the matrices. Demonstrating high specificity, sensitivity, and qPCR efficiency of about 91%. The limit of detection went as low as 2×10-5 of the stock virus dilution. The results were more comparable or higher by 10-fold to the column-based extraction method followed by aqueous phase two-step RT-qPCR (Kifaro et al., 2023).
Escaping the viral RNA purification step using commercially available kits while analyzing these complex samples without the use of a centrifuge for debris removal means the technology carries a high potential for onsite application. It is assumed that the presence of nanopores in the microparticles plays a role in protecting the enzymatic activity for viral RNA capture by preventing the high debris content from reaching the inside the particles. Furthermore, this technology will highly reduce the need for a stable cold chain for sample and reagent storage, adding the advantage of this technology.
Reducing the chances for cross-contamination
As proof of concept, the protocol has cut down the number of sample processing steps to three. Reducing the chances for cross-contamination and sample pre-processing time. The cDNA pool stability for a longer duration is key for the stable and efficient downstream process, including qPCR and sequencing. But also opens room for extensive viral RNA capture at field conditions, which is extremely important to speed up surveillance activities of infectious diseases.
The work in progress is to validate the technology using natural field samples.
References
- Bataille et al. (2019). Optimization and evaluation of a non- invasive tool for peste des petits ruminants surveillance and control. Sci Rep, 9(1), 4742.
- Dhumpa et al. (2011). Rapid detection of avian influenza virus in chicken fecal samples by immunomagnetic capture reverse transcriptase-polymerase chain reaction assay. Diagn Microbiol Infect Dis, 69(3), 258-265.
- Kifaro et al. (2023). Microparticles as Viral RNA Carriers from Stool for Stable and Sensitive Surveillance. Diagnostics. 13, 261.
- Prickett et al. (2008). Oral-fluid samples for surveillance of commercial growing pigs for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 infections. Journal of Swine Health and Production, 16(2), 86-91
Author’s acknowledgments
This article is based on a study conducted by Emmanuel Kifaro as part of his Ph.D. program at the Sokoine University of Agriculture and the Korea Institute of Science and Technology under the support of the Regional Scholarship and Innovation Fund (RSIF) of the Partnership for skills in Applied Sciences, Engineering and Technology (PASET) to SACIDS Foundation for One Health.. The author would like to thank Prof. Sang Kyung Kim, Dr. Seungwon Jung, and Ms. Mi Jung Kim for their tireless mentorship, and assistance, which aided in the completion of this study.

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.