Charles W. Carter, Jr, Department of Biochemistry and Biophysics, University of North Carolina Chapel Hill, reviews the ways that recent research in Structural Biology, Biochemistry, Molecular Biology, and Phylogenetics have opened the origins of genetic coding to experimental study and their important implications
Structural Biology is the study of the 3D arrangements of atoms in biological molecules. It is an immensely rich source of information that has continually transformed how we think of ourselves. Knowing a structure adds a new level of reality to an entire range of mechanistic models. The double helical structure of DNA likely brought the most fundamental overhaul of our perspective. It showed that units we had called “genes” have a structure that makes the idea of a “heritable blueprint” unmistakably self-explanatory.
Reading genes
Dennis Noble argues in a review of Phillip Ball’s new book, How Life Works (1), that “blueprint” is a “lazy metaphor”. (2) Understanding that the reality is indeed much more interesting is crucial because of how it informs policy. One of the ways to see his argument is to recognize how much is missing from the blueprint metaphor. The essence of the metaphor is that blueprints must be read out. Reading genes out has two implications that tend to be ignored.
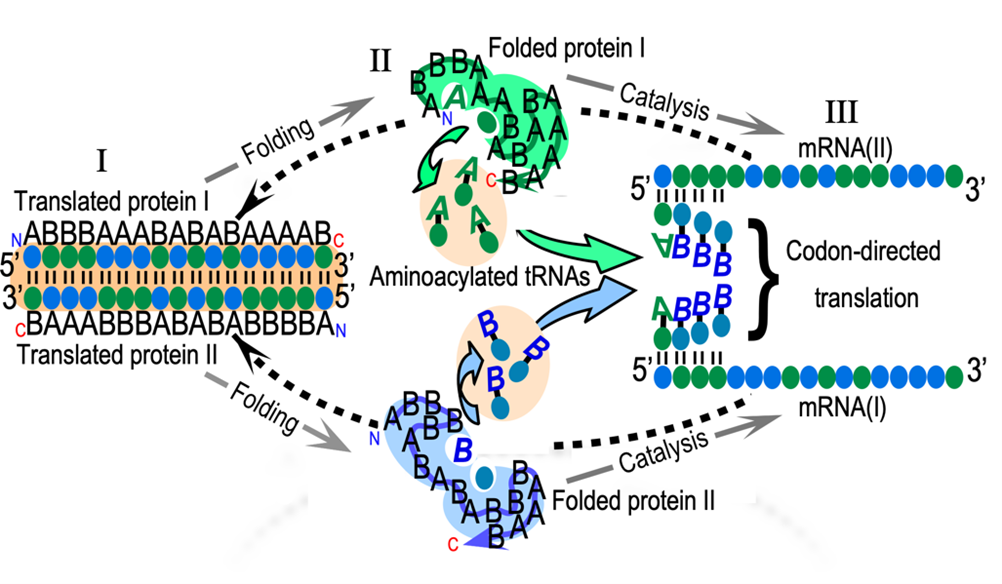
First, it implies a reflexive symbolic translation from one chemical language to another. That readout is done by a set of proteins, called aminoacyl-tRNA synthetases (AARS). AARS distinguish between 20 kinds of amino acids and 61 kinds of transfer (t)RNAs. When they bind both correctly, they form a chemical bond between them (Fig. 1). (3) That bond cements the translation of the code by connecting amino acids to RNAs containing the right symbols (called “codons”). The search for the origin of translation will succeed when we can describe the earliest AARS*tRNA “cognate pairs” and the rules by which the AARS recognized their two kinds of substrate. (4)
Second, translation creates protein products that form the core networks that power the cell. Nearly all of biology, both known and unknown, flows from the complex, often reflexive interactions between the elements of those networks. Protein elements – enzymes, motors, receptors, and regulatory proteins – amplify the functions of their genes by an immense factor, estimated to be 109-fold. (5,6)
Aminoacyl-tRNA synthetases (AARS)
Our quest for how Nature assembled the first AARS•tRNA cognate pairs has tried to adhere to the tenet that two things must have been true of ancestral AARS.
- i. They must have been functional in the sense that they could catalyze one or both of the chemical reactions necessary to assemble proteins in a templated fashion. This means that we must be able to demonstrate those functionalities in the laboratory. (7)
- ii. They must have had a strong phylogenetic connection to the most highly conserved structures across each family.
Experimental Biochemistry is the only means we have to assess functionality. Phylogenetics is the only record we have of what sequences might have survived a nearly random ancestry. The survival of those sequences and their ancestral functionality are clearly interdependent. Structural Biology played a key role in bringing us as far as we’ve come. My four previous segments (3,4,7,8) tell much of that story so far. Here, I use Fig. 1 to summarize where I think the field has come, and to outline how far we have yet to go.
Aligning the 3D atomic coordinates of all members of each AARS Class revealed that both superpositions show a sharp contrast between a common core, much smaller than the full-length enzymes, and a diverse, idiosyncratic collection of surface loops. However, those highly variable surface loops are inserted into the same places within the cores.
Moreover, the core-loop junctions can be replaced by a single peptide bond. These aspects of AARS molecular anatomy pointed us directly at the structural cores. It was conceptually straightforward to construct genes for the cores themselves, and only moderately difficult to purify them and show that they retained most of the catalytic proficiency of their full-length (putative) descendants. That path has thus far given us four AARS urzymes, two from each Class, that exhibit more or less complementary amino acid specificities. (9)
Along the way, we also discovered ways to tease out details of how ancestral AARS recognized their cognate RNA substrates by an operational code. (4) We related those specificities and the recognition of Class I and II amino acid substrates to projections of base- pairing between ancestral genes into the proteome. (3) Not a bad start. But these are really only starters. They only set the stage for the main tasks that remain to be taken on.
Ancestral gene sequences
The ultimate puzzle is how Nature built a set of protein decoders that could enforce the coding rules by which they, themselves, were assembled. That task is outlined in the details in (Fig 1). It entails genes written with an alphabet with as few as two distinct kinds of amino acids. The translated products of those genes had to fold into 3D structures whose catalytic apparatus, amino acid, and RNA substrate recognition could then impose the coding rules required to read their own gene sequences.
A critical missing piece is to strengthen the computer algorithms used to deduce ancestral sequences. These are the province of phylogenetics. (10,11) We analyze amino acid sequence alignments from many contemporary genes for positions where they differ and then estimate from the distributions of different side chains at those positions which amino acids the likely common ancestor used at that position. The biochemical tools we have developed should then provide the experimental platform to characterize those ancestral sequences. (7)
References
- P. Ball, How Life Works: A User’s Guide to the New Biology, University of Chicago, 2023.
- D. Noble, Nature, 2024, 626, 254-255.
- C.W. Carter, Jr., OpenAccessGovernment 2023, April, 54-55.
- C.W. Carter, Jr., OpenAccessGovernment, 2024, 41, 228-229.
- C. W. Carter, Jr and P. R. Wills, Molecular Biology and Evolution, 2018, 35, 269-286.
- P. R. Wills, Phil. Trans. R. Soc. A, 2016, A374, 20150016.
- C.W. Carter, Jr., OpenAccessGovernment 2023, July, 272-273.
- C. W. Carter, Jr., OpenAccessGovernment, 2023, October, 256-256.
- C. W. Carter, Jr., MDPI Life, 2024, 14, 199.
- J. Douglas, R. Bouckaert, C. W. Carter, Jr. and P. Wills, Nucleic Acids Research, 2024, 52,, 558–571.
- C. W. Carter, Jr., A. Popinga, R. Bouckaert and P. R. Wills, International Journal of Molecular Sciences, 2022, 23, 1520.

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.