Roksana Riddle and Christopher H. Contag from Michigan State University discuss the concept of endosymbiosis, how it has evolved, and present strategies to engineering endosymbionts and their applications in developing innovative therapies
What is endosymbiosis?
Endosymbiogenesisis is the emergence of mutually beneficial relationships between cells in which one lives inside the other to become a holobiont. Endosymbiosis has occurred naturally when intracellular parasites, dependent on host health, begin providing benefits to that host. While this jump may seem like an impossible change from harm to benefit, a study of five obligate intracellular parasites showed a loss of gene redundancy, elimination of metabolic genes, and, most importantly, diminished defense mechanisms when compared to free-living bacteria such as E. coli (Sakharkar et al., 2004). If a parasite is host-dependent, then any defensive genes that harm the host further would alert the immune system, resulting in the destruction of the partnership.
Redundant genes increase the metabolic burden of genome maintenance and protein production, which subsequently increases the metabolic burden on the host. A stressed host can starve, or signs of unhealth can lead to immune destruction of the infected cell. These observations indicate that the best candidates for endosymbionts have a reduced genome size and increased specialization. They must also have the ability to enter the cell and perhaps exist without causing damage or immune activation, and there must be mutual dependence on holobiont survival and matched rates of cell division (Fig. 1).
Endosymbiont relationships are thought to be the foundation upon which multicellular eukaryotic life was built, according to the endosymbiont theory of mitochondrial evolution (Fig. 2).
This theory postulates that eukaryotes could not evolve the genetic complexity to become multicellular without the ATP generated by the mitochondria (Lane et al., 2010). At some point, the ancestral alphaproteobacterial parasite, which benefitted from the complex macromolecules produced in the holobiont, had the orientation of its ADP/ATP translocator swap from the ancestral copy and began benefiting the holobiont energetically by pumping ATP into the host cell (Martin and Zimorski. 2015). Over time, the mitochondria specialized in energy production, allowing it to maintain a smaller genome containing only 13 protein-coding regions in human cells, including those for oxidative phosphorylation and the ADP/ATP translocase. This original endosymbiosis is mirrored in other extant examples, and this path serves as a guide for the development of innovative endosymbiont-based therapies.
Animal endosymbionts
One extant example of an evolved endosymbiosis is the relationship between corals and their Zooanthae algae. This algal symbiont evolved from a parasite but now provides up to 95% of the coral’s energy needs in exchange for diverse metabolites and protection (Fournier, 2013). This gives corals the ability to support entire ecosystems in nutrient-poor but sunny shallows. Interestingly, Zooanthae can live as a free-floating species while the coral bleaches and dies without its energy partner; this symbiotic relationship has led to the coral having a much smaller genome (LaJeunesse, 2020).
Other examples of co-evolved intracellular bacterial parasites are Wolbachia and Mycobacterium leprae. Wolbachia is a bacterium that can protect its mosquito host from infections by viruses such as Dengue, Chikungunya, and Plasmodium by preventing the replication of its competitor (Luciano et al., 2009). However, Wolbachia can cause infertility in the mosquito hosts as part of its replication – this is an imperfect symbiosis.
Mycobacterium leprae causes liver regeneration in its armadillo host by reprogramming adult Schwan cells into progenitor stem-like cells that can differentiate into complex liver tissues (Hess et al., 2022). In an armadillo with a normally healthy liver, this is a problem and mainly serves to ensure the spread of the bacteria. This model is particularly attractive from a tissue engineering standpoint, considering one out of every 25 human deaths is from liver failure; a treatment could lie in engineered bacteria (Harshad et al., 2023). These symbiotic relationships are new and imperfect; the need for the symbiont to spread, despite the host not needing the symbiont, causes harm to the host.
Engineering endosymbionts
Engineering endosymbiotic relationships is an emerging research area with obvious clinical uses and the potential to become a whole new field of medicine. Despite several clinically approved bacterial therapies, endosymbiont therapies are still in their infancy. Understanding the endosymbiont relationship such that it can be disrupted has led to a breakthrough in how Filariae worms that cause blindness are treated in Africa; despite efforts to kill the worms proving ineffective, targeting the worm’s Wolbachia endosymbionts with antibiotics renders the worms infertile preventing their long-term spread (Hoerauf et al., 2000). In an attempt to engineer lasting endosymbiosis, oxidative phosphorylation was disrupted in yeast mitochondria, making them dependent on the ATP produced by glycolysis; meanwhile, E. coli was given the ADP/ATP translocase so that it could provide energy to the yeast when glycolysis was not available creating a lasting holobiont population (Mehta et al., 2018). A key part of this model’s success is the E. coli’s dependence on its holobiont for thiamin diphosphate, which makes it unable to live without a healthy host. Additionally, the yeast and E. coli doubling times are similar: 90 and 30 minutes in optimal media, respectively. Yeast cells can be simple holobionts since they are unicellular and lack immune systems, but they do not help address all the challenges presented by multicellular holobionts.
Potential medical applications
Bacillus subtilis cells phagocytized by macrophages were engineered to secrete Listeriolysin O to escape the phagosome (Madsen et al., 2022). Once in the cytosol, the bacteria were induced to secrete mammalian transcription factors that activate genes in immune pathways, demonstrating the potential of engineered endosymbionts to redirect immune cells to destroy tumors (Madsen et al., 2022). Unfortunately, these results decreased when measured at 48 hours due to a combination of destruction of the endosymbiont through autophagy and B. subtilis’ rapid division time leading to death of the holobiont. Learning from these experiments, future engineered endosymbionts need to have small genomes, be slow- growing, depend on host survival, and be able to evade immune destruction.
JCVI-Syn3B
JCVI-Syn3B was created by the J. Craig Venter Institute (JCVI) as a model to study minimal genome life and to study genes isolated from complex systems. It was constructed by starting from an already small genome organism, Mycoplasma mycoides capri, and sequentially removing genes from a fully synthesized genome until only those necessary for survival, morphology, and growth remained (Breuer et al., 2019). Still far from the mitochondrial 13 protein ideal minimal genome, Syn3B’s 452 protein-coding regions make the synthetic bacterium a useful chassis for creating endosymbionts. A reason mitochondria cannot easily be engineered into cell fate-controlling endosymbionts is that they cannot replicate autonomously. Most of the required mitochondrial proteins are encoded in the nucleus. Mycoplasma spp are infamous as cell culture contaminants due to their small size, persistence, and lack of immune recognition.
Mycoplasma do not have cell walls. Syn3B additionally lacks many surface proteins or sugars, causing it to nearly appear a simple liposome to immune cells with little or no reactivity. The lack of an immune response may illicit fear about the potential for engineered Syn3B endosymbionts to become pathogenic, but the minimal genome size addresses many of these concerns. Without redundant genes that are free to mutate, it is unlikely that there would be a significant gain of function in this bacterium. Also, Syn3B lacks the genes encoding attachment proteins, making acquisition of new host cell tropism or binding unlikely. While not as close as the yeast and the E. coli engineered symbiosis, the slower doubling time of Syn3B, two hours, makes it less likely to cause problems when delivering proteins to human cells, which have a varying but generalizable doubling time of around 24 hours. Syn3B lack the genes that would limit its success as an endosymbiont, but it retains the SecYEG secretion complex allowing it to deliver proteins to the cytoplasm of a future holobiont with mammalian transcription factors that alter cell fates (Pedreira et al., 2022).
Challenges
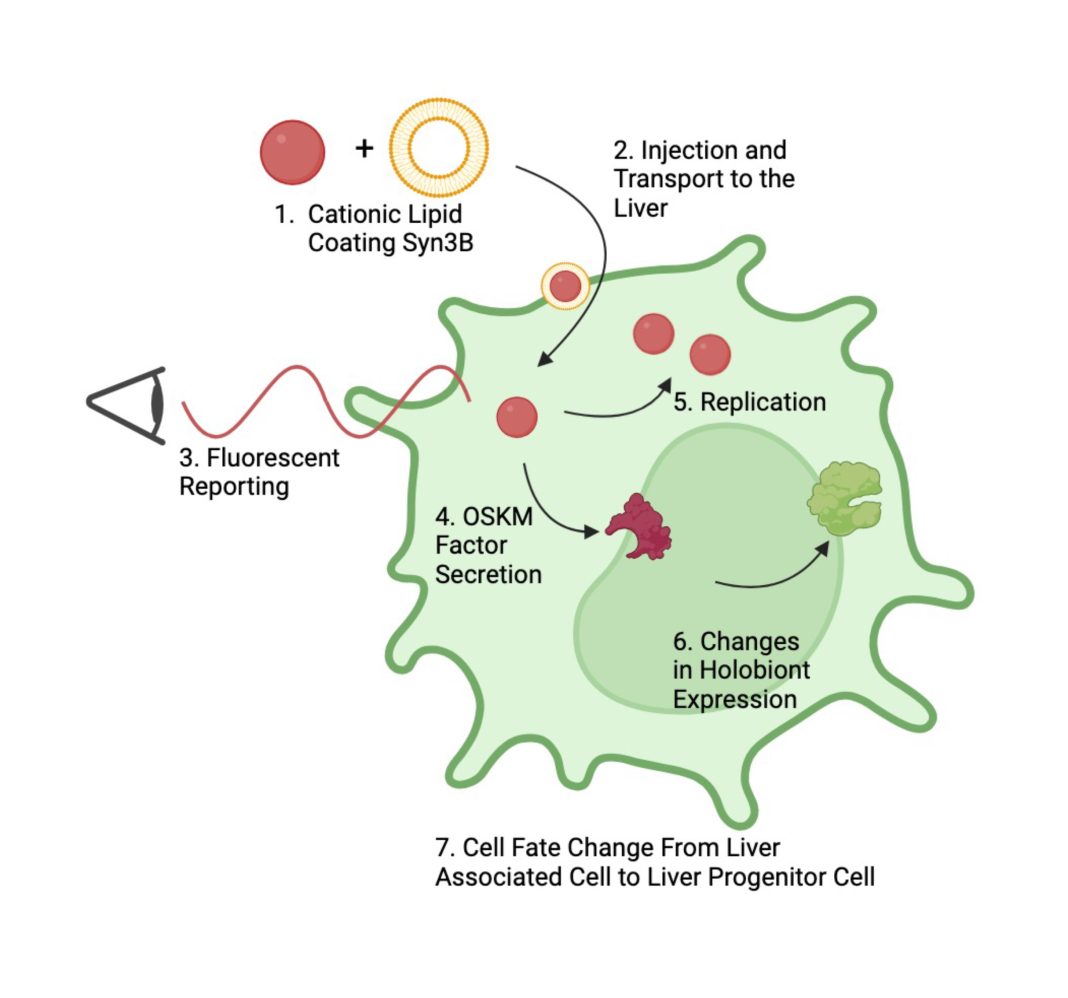
One step to overcome in developing Syn3B-based endosymbionts is targeted entry into the cytoplasm of cells in specific tissues. Tumor-associated macrophages have been an attractive target because of the ability of the endosymbiont to be phagocytized by activated macrophages. Without recognition by macrophages and no attachment proteins, Syn3B is unlikely to be taken up. One potential method to overcome this challenge is the use of targeting cationic lipid coatings. While not in the ideal size range of deliverable therapeutics, Syn3B is a 400nm in diameter sphere; it has a negatively charged lipid cell membrane, making it compatible with cationic lipid coatings for cellular uptake. In a study by Dillard et al. 2021 lipid-coated nanoparticles (LNP) were formulated with a ‘SORT’ lipid that picked up a protein corona of specific blood proteins, which directed LNPs to either the liver, spleen, or lung tissue-based on the SORT molecule used. This approach would also reduce the toxicity of coated Sny3B because, after entry, the coating would be removed, eliminating the possibility of reinfection. Other potential aspects of Syn3B to consider are its slow metabolism and low protein output; an endosymbiont that adds function to its host needs to persist and deliver enough protein to create a phenotypic change.
Minimal genome endosymbionts for tissue and organ regeneration
Engineered endosymbionts may be ideal for a therapeutic strategy for tissue regeneration. Four transcription factors for cellular reprogramming, Oct4, Sox2, Klf6, and c-Myc (Takahashi et al., 2007), could be delivered by endosymbionts to induce pluripotency, and other transcription factors subsequently guide tissue regeneration. Pluripotent cells can differentiate into almost any other cell type and, with proper signals, can become complex organ tissue, as demonstrated by the armadillo model. An example of how stem cells differentiating into the liver can provide health benefits is a study by Nagamoto et al. 2016 finding that after carbon tetrachloride damage, mouse survival rate jumped from 16.7% to 63.2% after engraftment of a sheet of induced pluripotent stem cell-derived hepatocyte-like cells. This incredible result required the inducement of pluripotent stem cells over two weeks, the 11-day differentiation into hepatoblast-like cells (liver stem cells), and a 46-day process of assembly into a sheet of liver tissue ready for surgical transplant. These observations would suggest that a treatment comprised of liver-targeted, SORT-coated, engineered Syn3B that secrete OSKM factors could lead to directed liver regeneration.
To summarize, Syn3B is an ideal model for engineering endosymbiosis for reprogramming cell fates and may have a variety of clinical applications. With its ability to harmlessly enter a cell, sustain protein delivery without immune activation, and its low energy costs, Syn3B is well aligned with evolved examples of endosymbiosis. The minimal genome organism, Syn3B, is an ideal endosymbiont chassis that will lead to innovative new therapies for guided replacement of diseased or damaged tissues.
References
- Lane, N., Martin, W. The energetics of genome complexity. Nature 467, 929–934 (2010). https://doi.org/10.1038/nature09486
- Martin, WF., Garg, S., Zimorski, V. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B Biol Sci (2015). Sep 26;370(1678):20140330. doi: https://doi.org/10.1098/rstb.2014.0330. PMID: 26323761; PMCID: PMC4571569.
- Fournier, A. The story of symbiosis with zooxanthellae, or how they enable their host to thrive in a nutrient-poor environment. Master BioSciences, Biology department, Ecole Normale Supérieure, Lyon. CRIOBE USR 3278 CNRS EPHE Papetoai, Moorea. (2013) https://biologie.ens-lyon.fr/ressources/bibliographies/pdf/m1-12-13-biosci-reviews-fournier-a-2c-m.pdf?lang=fr
- LaJeunesse, T. Zooxanthellae. Current Biology 30, 19 (2020) https://www.cell.com/current-biology/fulltext/S0960-9822(20)30428-0
- Luciano, M., Iturbe-Ormaetxe, I., Jeffery, J. A Wolbachia Symbiont in Aedes aegypti Limits Infection with Dengue, Chikungunya, and Plasmodium Cell, Volume 139, Issue 7, 1268 – 1278 (2009). https://www.cell.com/fulltext/S0092-8674(09)01500-1
- Hess S., Kendall TJ., Pena, M., Yamane, K., Soong, D., Adams, L., Truman, R., Rambukkana, A. In vivo, partial reprogramming by bacteria promotes adult liver organ growth without fibrosis and tumorigenesis. Cell Rep Med (2022). Nov 15;3(11):100820. doi: https://doi.org/10.1016/j.xcrm.2022.100820. PMID: 36384103; PMCID: PMC9729881.
- Hoerauf, A., Volkmann, L., Hamelmann, C., Adjei, O., Autenrieth, I. B., Fleischer, B., & Büttner, D. W. Endosymbiotic bacteria in worms as targets for a novel chemotherapy in filariasis. The Lancet, 355(9211), 1242–1243 (2000). doi:https://doi.org/10.1016/s0140-6736(00)02095-x
- Mehta AP, Supekova L, Chen JH, Pestonjamasp K, Webster P, Ko Y, Henderson SC, McDermott G, Supek F, Schultz PG. Engineering yeast endosymbionts as a step toward the evolution of mitochondria. Proc Natl Acad Sci U S A (2018). Nov 13;115(46):11796-11801. doi: https://doi.org/10.1073/pnas.1813143115. Epub 2018 Oct 29. PMID: 30373839; PMCID: PMC6243291.
- Madsen, C.S., Makela, A.V., Greeson, E.M. et al. Engineered endosymbionts that alter mammalian cell surface marker, cytokine, and chemokine expression. Commun Biol 5, 888 (2022). https://doi.org/10.1038/s42003-022-03851-6
- Breuer, M., Earnest, T., Merryman, C., Wise, K., Sun, L., Lynott, M., Hutchison, C., Smith, H., Lapek, J., Gonzalez, D., de Crécy-Lagard, V., Haas, D., Hanson, A., Labhsetwar, P., Glass, J., Luthey-Schulten, Z. (2019) Essential metabolism for a minimal cell eLife 8:e36842. https://doi.org/10.7554/eLife.36842
- Pedreira T., Elfmann C., Singh N., Stülke J. SynWiki: Functional annotation of the first artificial organism Mycoplasma mycoides JCVI-syn3A Protein Sci (2022). Jan;31(1):54-62. doi: https://doi.org/10.1002/pro.4179.
- Dilliard, SA., Cheng, Q,. Siegwart, DJ. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc Natl Acad Sci U S A (2021). Dec 28;118(52):e2109256118. doi: https://doi.org/10.1073/pnas.2109256118. PMID: 34933999; PMCID: PMC8719871.
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell (2007). Nov 30;131(5):861-72. doi: https://doi.org/10.1016/j.cell.2007.11.019. PMID: 18035408.
- Transplantation of a human iPSC-derived hepatocyte sheet increases survival in mice with acute liver failure Nagamoto, Yasuhito et al. Journal of Hepatology, Volume 64, Issue 5, 1068 – 1075
- Harshad, D., et al. Global burden of liver disease: 2023 update Journal of Hepatology, Volume 79, Issue 2, 516 – 537 (2023).
- Sakharkar KR, Dhar PK, Chow VTK. Genome reduction in prokaryotic obligatory intracellular parasites of humans: a comparative analysis. Int J Syst Evol Microbiol (2004). Nov;54(Pt 6):1937-1941. doi: https://doi.org/10.1099/ijs.0.63090-0. PMID: 15545414.