Serge L. Beaucage investigates thermolabile protecting groups for the amine functions of purine and pyrimidine deoxyribonucleosides for the development and implementation of synthetic DNA sequences as nucleic acid-based drugs
The high demand for nucleic acid-based drugs, earmarked for treating human diseases, requires improved chemical methods for rapid and efficient chemical synthesis of synthetic DNA and/or RNA sequences with protecting groups before being processed and manufactured in sufficient quantity and purity for clinical indications.
Given that the purity of synthetic DNA/RNA sequences is of the utmost importance for their intended purposes, sustained research efforts have been made.
Examining DNA sequences and so much more at the Beaucage lab
Over the years, researchers at the Beaucage lab have tried to improve the solid-phase chemical synthesis and solid-phase purification of DNA and/or RNA sequences.
Specifically, the thermolabile 4-methylthio -1-butyl and 2-(N-formyl-N-methyl) aminoethyl groups ([Cieślak et al.,] [https://pubs.acs.org/doi/10.1021/jo035861f]; [Grajkowski et al.,] [https://doi.org/ 10.1021/ol0156852]) for phosphate/thiophosphate protection of synthetic DNA sequences, are distinctive in terms of structural simplicity and coupling efficiency of their respective deoxyribonucleoside phosphoramidite derivatives.
DNA sequences have been shown to have several phosphate and thiophosphate protecting groups that can eventually be cleaved through an efficient intramolecular cyclodeesterification reaction.
This occurs within minutes up to hours when exposed to temperatures ranging from 37°C-90°C under near-neutral (pH 7.4) or mildly basic (pH 9) conditions in an aqueous buffer ([Ausin et al.,] [https://doi.org/10.1016/j.tet.2009.10.096]; [Grajkowski et al.,] [https://doi.org/ 10.1039/B9NJ00692C] ).
Thermolytic cleavage of protecting groups
Inspired by the implementation of the thermolabile 2-(N-formyl-N-methyl) aminoethyl phosphate/thiophosphate protecting group ([Grajkowski et al.,] [https://doi.org/10.1021/ol0156852]) in the solid-phase synthesis of model oligodeoxyribonucleotides, protection of the purine and pyrimidine amine functions of deoxyribonucleosides, as N, N-dimethyloxalamides, is reported herein along with the conditions used for thermolytic cleavage of these protecting groups.
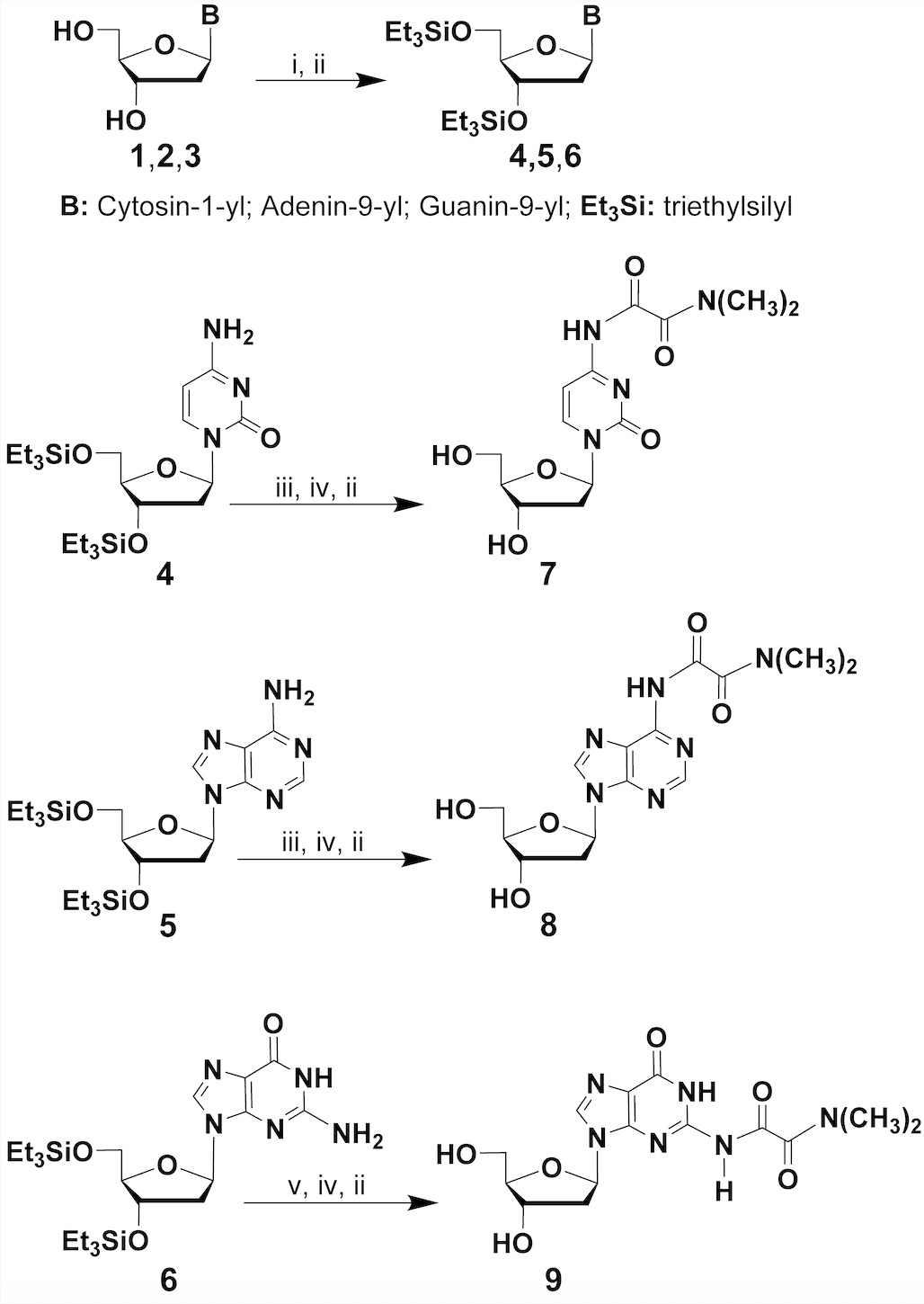
As presented in scheme 1, the hydroxyl functions of the native deoxyribonucleosides 1-3 are first protected as their respective triethylsilyl ethers 4-6. The exocyclic amine functions of 4-6 are then protected as N, N-dimethyloxalamides, under the conditions described in the caption of scheme 1, to provide the N-protected deoxyribonucleosides 7-9.
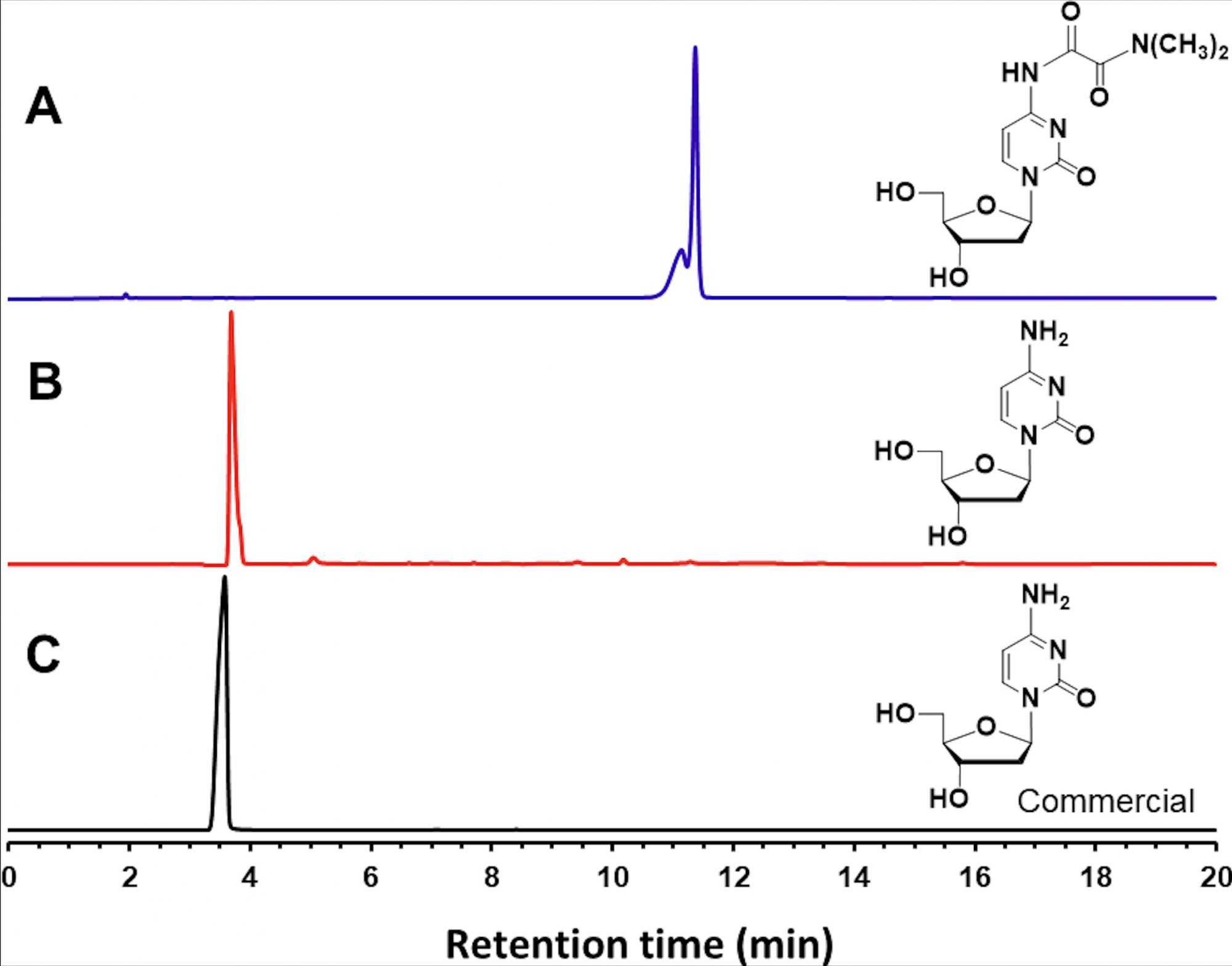
The thermostability of N, N– dimethyloxalamides, as amine-protecting groups, has been investigated. Indeed, heating 7 in a Trizma hydrochloride solution (pH 9) at 90°C over one hour has led to complete thermolytic cleavage of its amine- protecting group.
Furthermore, as illustrated in figure 1, the RP-HPLC profile of 7 demonstrates quantitative N4-deprotection to provide deoxycytidine compared to the RP-HPLC profile of a commercial deoxycytidine sample.
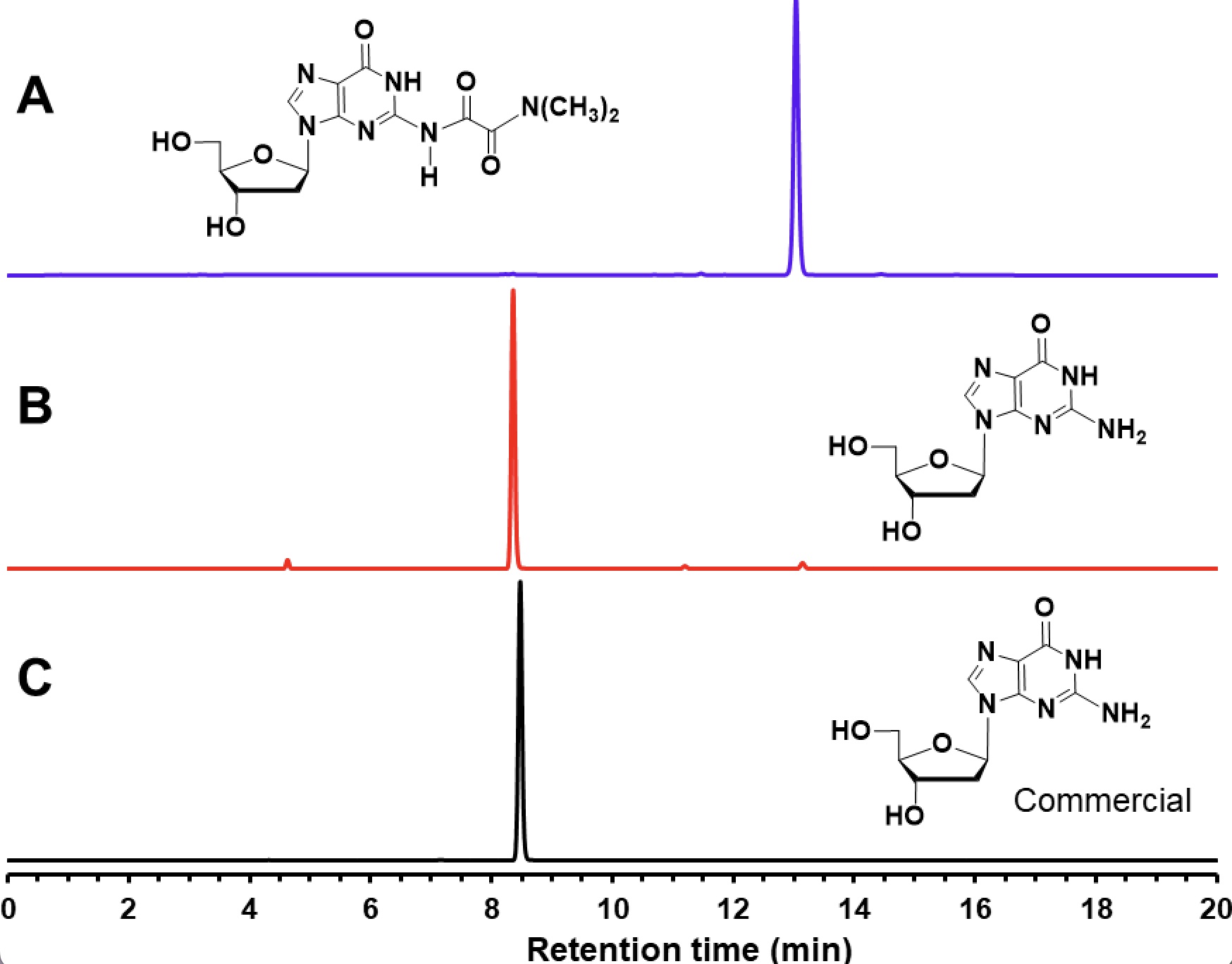
The RP-HPLC profile of 8 presented in figure 2 validates the quantitative N6– deprotection of 8, within one hour at 90°C in a Trizma hydrochloride solution (pH 9) to provide deoxyadenosine, when compared to the RP-HPLC profile of a commercial deoxyadenosine sample.
Like the chromatographic profiles supporting the facile thermolytic deprotection of the amine-protecting group from the deoxyribonucleosides 7 and 8, RP-HPLC analysis of N2-protected 9 also shows quantitative amine deprotection under essentially identical thermolytic conditions to those reported in the caption of figure 1.
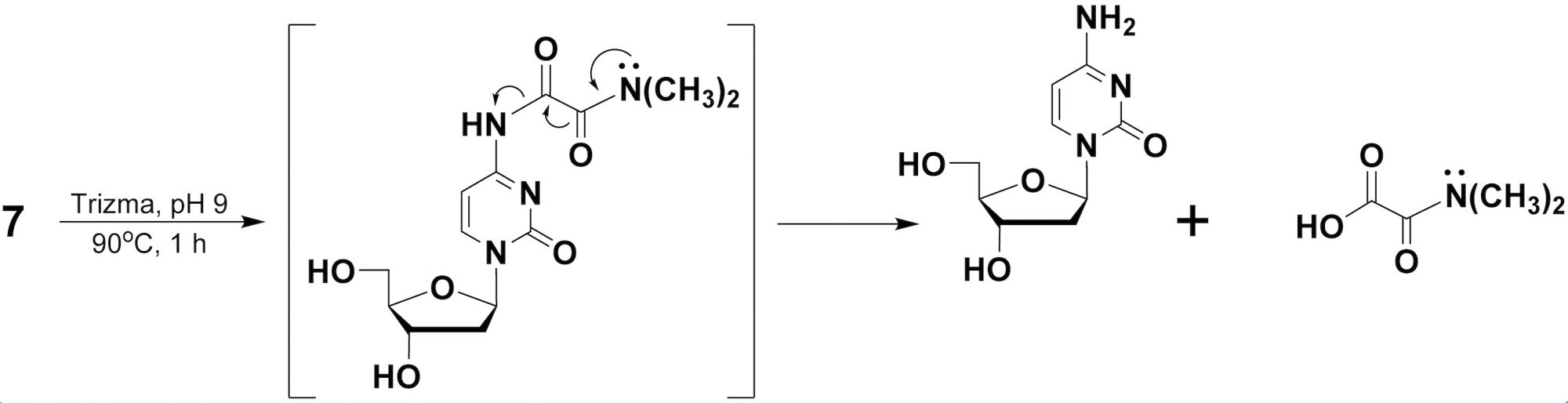
The mechanistic pathway for thermolytic deprotection of N, N- dimethyloxalamides, as amine protecting groups, is very likely to proceed through thermal hydrolysis and/or potentially through an intramolecular deamidation pathway, as proposed in scheme 2.
Protection of the amine function of deoxyribo- nucleosides and the application of N, N-dimethyloxalamide protecting groups
The outcome of the proposed strategy for the protection of the amine functions of purine and pyrimidine deoxyribonucleosides, and the facile thermolytic cleavage of N, N- dimethyloxalamides at pH 9 without the formation of DNA-modifying side products, strongly encourages the application of such protecting groups to the preparation of synthetic DNA and RNA sequences given the comparable chemical properties of the purine and pyrimidine amine functions of ribonucleosides to those of deoxyribonucleosides.

Thus, using thermolabile protecting groups for the amine functions of purine and pyrimidine deoxyribonucleosides or ribonucleosides can significantly facilitate the manufacture of pure, efficacious, and safe nucleic acid-based drugs for therapeutic applications.
Collaborators:
Sivakoteswara Rao Mandadapu Brian M. Cawrse

This work is licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International.


