Prof. Dr Marc Bramkamp explains how the latest science allow for increased understanding of the subcellular organisation of a bacterial cell
When we think about bacteria, we have a small round or rod-shaped organisms in mind that dwell as individual, planktonic bacterial cell without any sophisticated internal organisation. This common view was shaped by the fact that research on bacteria was focussing mainly on bacteria studied in test tubes during the last century. Since many biochemical pathways were discovered and analysed in bacteria, or more precisely in isolated bacterial extracts, the subtle feeling manifested that these organisms are nothing more than encapsulated reaction vessels in which biochemical reactions are spatially and temporally not well organised and driven merely by diffusion. However, we are currently in the middle of a revolution that changes the way we see (and understand) bacteria.
Bacteria are not individualists, but actively work together
We have learned that bacteria are by no means individualists, but able to act in concert, actively shape their environment (which in fact is also us), they communicate chemically and they share and distribute labour.
Thus, it is maybe less surprising that these activities are going hand-in-hand with cellular differentiation. This differentiation can occur at a metabolic, or biochemical, level and even on a morphological level. Bacteria might develop into a biofilm full of cell chains or cells may transform in dormant stages, such as spores that lie idle until favourable conditions occur.
How are nanometre-sized proteins organising micrometre sized cells?
For a long time, it was unclear how micrometre sized organisation such as the cell morphology was achieved by the action of nanometre-scale protein machineries. Better insights were prevented by lack of high-resolution imaging techniques. The main bottleneck was the resolution limit of visible light, as used in conventional wide-field microscopes. This physical limit allows the precise determination of objects only down to about 200-300 nm. The prototypical bacterial cell is less than 1000 nm in width.
Therefore, spatial information is limited when subcellular localisations of biomolecules, such as protein or nucleic acids are addressed. Furthermore, it needs fluorescent dyes to label individual molecules (proteins or nucleic acids) to allow their visualisation within the dense mixture of material in a cell.
New microscopy techniques are available now
The last two decades have given us new tools that allow microscopic imaging of subcellular structures in bacterial cells. A variety of different fluorescent proteins have been developed that can be used for translational fusions to identify protein localisation. Sophisticated dyes such as photo-switchable proteins can be used elegantly to track protein dynamics in living (bacterial) cells.
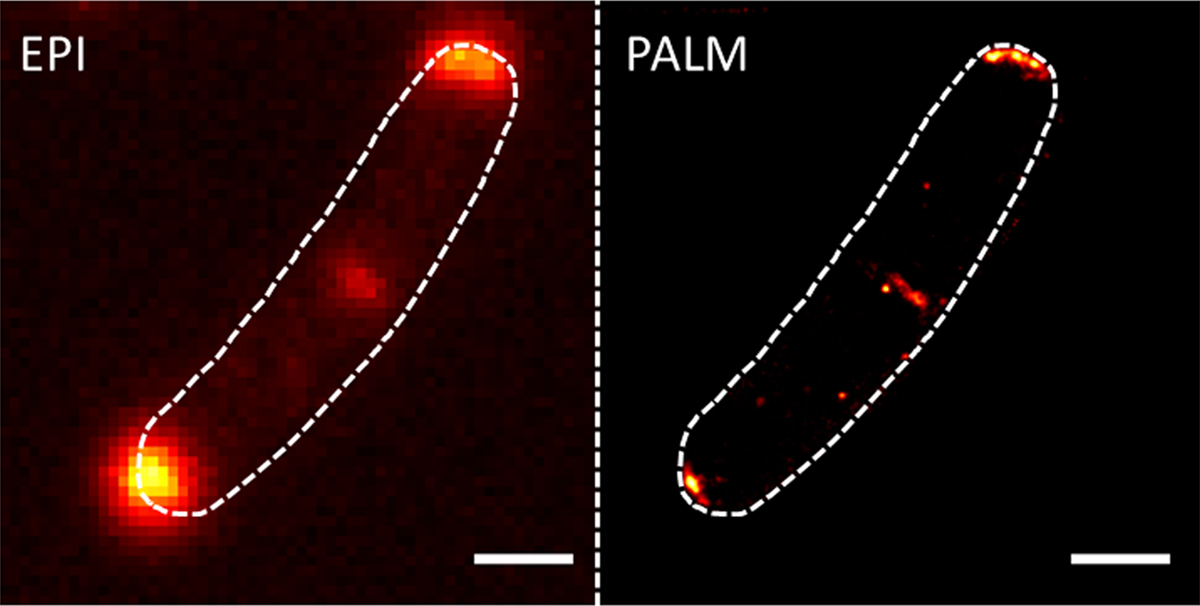
New microscopic techniques that circumvent the resolution limit of light also greatly helped in gaining deeper insights into the organisation of cells. My laboratory uses photo-activated localisation microscopy (PALM), a technique that allows unrivalled precision in protein localisation down to 20 nm in cells (Figure 1). We develop and optimise dual colour PALM imaging by combining various fluorophores such as mNeonGreen and photoactivatable-mCherry.
The bacterial plasma membrane is spatially organised in domains
With these modern techniques in the toolbox, my research team tries to answer several central questions in bacterial cell biology. The main topic is the organisation of the bacterial plasma membrane. This phospholipid and protein made cellular envelope is highly dynamic, but we start to appreciate that certain proteins and lipids cluster in defined regions in which they function optimally. Cells have evolved proteins that help to create membrane heterogeneity, to allow the existence of distinct reaction areas in which transport or signalling events are enhanced. Proteins involved in the formation or maintenance of the membrane domains are so-called flotillins, a class of proteins that are conserved from bacteria to man. PALM images show how that the flotillins indeed form a distinct cluster in the membrane.
We are further interested how the cell keeps its membrane intact. This is an important aspect since the cell membrane is a classical target of many antimicrobial compounds. We identified a bacterial dynamin that acts in membrane surveillance. Upon membrane rupture, caused by antimicrobial compounds or phages, the bacterial dynamin clusters at the damaged sites and seals the membrane via lipid fusion. These are just two examples were bacteria use proteins that were long thought to be evolutionary achievements of eukaryotic cells.
Bacterial cell wall synthesis is spatially highly organised
Bacterial morphogenesis and cytokinesis are accomplished by highly complex protein machinery that catalyses the synthesis of the bacterial cell wall. The peptidoglycan (or murein) is a unique structure to bacterial cells and therefore a prime target of many potent antibiotics.
Although most bacteria have a reasonably simple morphology in being round (cocci) or rods (bacilli), we observe a remarkable difference in the spatial organisation of the cell wall synthesis machinery. Many rod-shaped bacteria position their cell wall synthetic complexes along the lateral axis of the cell. This is true for Gram-negative bacteria such as Escherichia coli, but also for the Gram-positive organism Bacillus subtilis.
Interestingly, actinobacteria, among them the rod-shaped Corynebacteria and Mycobacteria have polar localised cell wall synthesis complexes. Therefore, these species grow from their ends. We have chosen Corynebacterium glutamicum as a model organism to understand the spatiotemporal organisation in these cells for some simple reasons. C. glutamicum is an important organism in biotechnology and is used to produce amino acids and other fine chemicals.
Furthermore, C. glutamicum shares its complex, multi-layered cell envelope with known pathogens such as Mycobacterium tuberculosis, the causative agent of TB. Our work revealed that apical cell growth is governed by cytoskeletal hub proteins that couple cell elongation and chromosome organisation. This close interaction between chromosome organisation and elongation growth may be an Achilles heel that hints to new antibiotic targets to combat drug-resistant Mycobacteria.
These research topics give a glimpse into how basic research subcellular organisation in bacteria may lead to applied research. There is increasing effort worldwide and within Germany to address the subcellular compartmentalisation of bacterial cells.
Therefore, we are grateful that our work on spatiotemporal organisation of bacteria is supported by grants from the German Federal Ministry of Education and Research (BMBF BMBF: 031A302 e:Bio-Modul II: 0.6 plus) and the Deutsche Forschungsgemeinschaft (DFG TRR174 & BR2915/6-1).
References:
Böhm K, Meyer F, Rhomberg A, Kalinowski J, Donovan C, Bramkamp M. (2017) Novel Chromosome Organisation Pattern in Actinomycetales-Overlapping Replication Cycles Combined with Diploidy. MBio. 8(3). pii: e00511-17. doi: 10.1128/mBio.00511-17.
Schubert K, Sieger B, Meyer F, Giacomelli G, Böhm K, Rieblinger A, Lindenthal L, Sachs N, Wanner G, Bramkamp M. (2017) The anti-tuberculosis drug ethambutol selectively blocks apical growth in CMN group bacteria. MBio 8(1). pii: e02213-16. doi: 10.1128/mBio.02213-16.
Baumgart M, Schubert K, Bramkamp M, Frunzke J. (2016) Impact of LytR-CpsA-Psr Proteins on Cell Wall Biosynthesis in Corynebacterium glutamicum. J Bacteriol. 198, 3045-3059.
Donovan C, Bramkamp M. (2014) Cell division in Corynebacterineae. Front Microbiol. 5:132. doi: 10.3389/fmicb.2014.00132.
Sieger B, Schubert K, Donovan C, Bramkamp M. (2013) The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol Microbiol. 90, 966-982.
Donovan C, Schauss A, Krämer R, Bramkamp, M. (2013) Chromosome segregation impacts on cell growth and division site selection in Corynebacterium glutamicum. PLoS One. 8, e55078. doi: 10.1371/journal.pone.0055078.
Donovan, C., Sieger, B., Krämer, R., and Bramkamp, M. (2012). A synthetic Escherichia coli system identifies a conserved origin tethering factor in Actinobacteria., Mol Microbiol 84, 105-116.
Please note: this is a commercial profile
Prof. Dr Marc Bramkamp
Ludwig-Maximilians-Universität München,
Fakultät Biologie
Großhaderner Straße 2-4
82152 Planegg-Martinsried, Germany
Tel: +49 (0)89 2180 74611
Fax: +49 (0)89 2180 74621