Frederique Lisacek from SIB Swiss Institute of Bioinformatics, ponders if we are getting closer to reproducing breast milk in baby formula
Milk content
It has been known for a while that breast milk plays an immuno-protective role for a newborn to prevent infectious diseases. This protection depends on a broad range of active components, including immunoglobulins (antibodies), immunocompetent cells, fatty acids, and glycoproteins. (1) Less known is the importance of lactose-derived molecules.
Lactose is a type of sugar. Sugar is synonymous with carbohydrates and takes various forms. The simplest sugars are the monosaccharides, for example, glucose or fructose, which are familiar terms in common use. For chemists and biochemists, assemblies of monosaccharides make up what are called oligosaccharides and polysaccharides. Lactose is a disaccharide made of glucose and a galactose molecule.
A diversity of oligosaccharides, designated as HMOs for Human Milk Oligosaccharides, is found in breast milk. These complex sugar molecules are almost all stemming from free lactose. HMOs are the third most abundant constituent of milk, after lactose and lipids, with total mean concentrations ranging from 4–30 g/L. Although indigestible by the neonate, it has been demonstrated that HMOs provide several benefits spanning antimicrobial action, protection from viral pathogens, promotion of healthy gut microbiota and the development of the immune and nervous systems.
Protective features
As explained in previous articles (1), the cell surface is coated with a wide variety of sugar molecules attached to proteins and lipids (glycoconjugates) and as such, mediate communication between cells. To perform their protective role, HMOs act as decoys to block the attachment of viral, bacterial or parasite pathogens to the sugar coat of a target cell surface.
HMOs range from three to over thirty monosaccharides in size and differ in composition from those of any other mammals. They are synthesised in the mammary gland and produced during lactation. Enzymes “build” HMOs from lactose by assembling monosaccharides in a stepwise process. The total content of breast milk in HMOs varies depending on individuals (genetic origin) and is impacted by diet and other environmental conditions. So far, between two and three hundred have been detected, but no one can ascertain what is the upper limit. Nonetheless, a feature unique to human milk is the heterogeneity of its HMO population, when compared to kinds of milk from other species, such as bovine, which are lower in both quantity and diversity. By presenting a wide variety of decoys, HMOs not only protect against pathogens, but simultaneously promote the colonisation of beneficial gut flora of Bifidobacterial species, thereby correlating HMO activity with balancing processes of the microbiome.
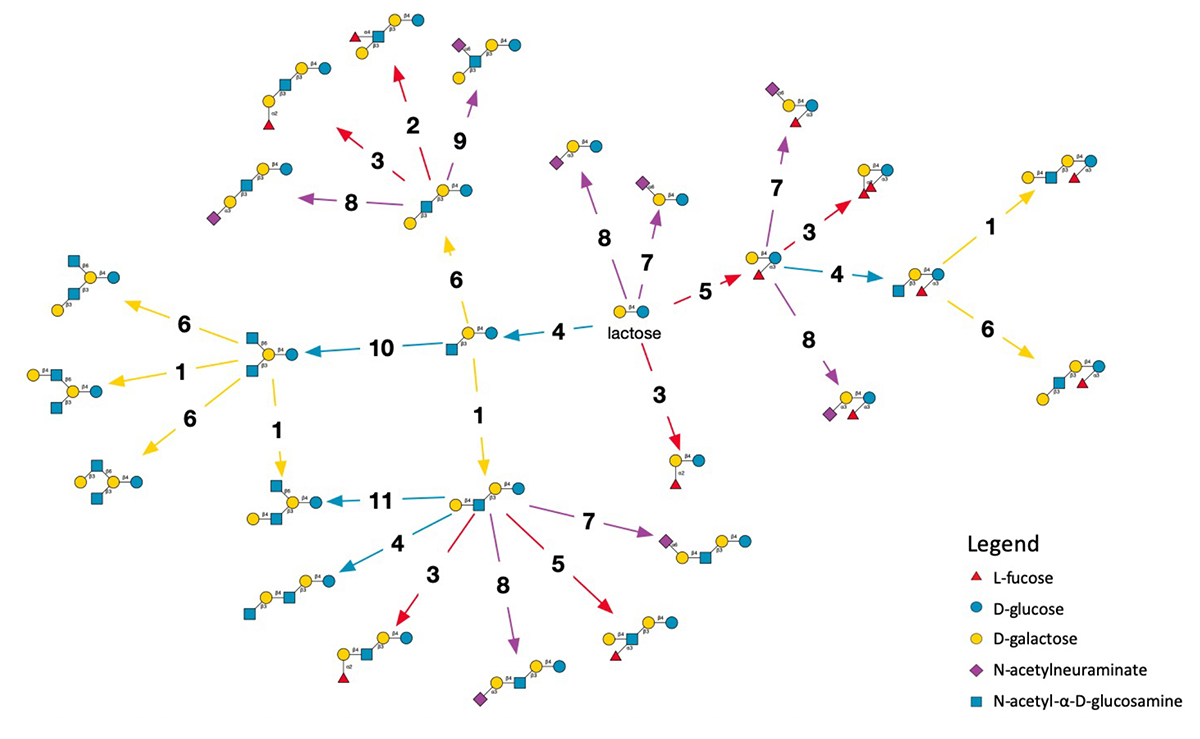
In this context, there has been much interest towards the synthesis of HMOs industrially, for use as probiotics as well as to improve baby formula. Although little is known as to the actual enzyme activities expressed in the lactating mammary epithelium, some insights can be gleaned from analysis of the HMO structures already characterised. In our research, we identified a subset of eleven enzyme activities that can account for over 200 distinct HMOs isolated from human milk. We have used this knowledge to construct an enzymatic reaction network that simulates HMO biosynthesis. This model provides a “chassis” for the most common HMOs found in lactating mothers.
Modelling and simulation
The smallest model network employing all activities of the eleven enzymes is shown in Figure 1. It represents monosaccharides in the Symbol Nomenclature For Glycans (SNFG) (2) that is widely adopted in biology to simplify the chemical notation that includes every atom with the bonds linking them. With SNFG symbols, glucose is a blue disc, galactose a yellow disc, therefore, lactose is pictured as a yellow and a blue disc tied together. The figure shows a reaction network generated by three iterations of the simulator, starting from lactose (centre). Enzyme reactions are represented as arrows leading from a precursor to a product. They are numbered from one to 11 and coloured according to the type of monosaccharide transferred.
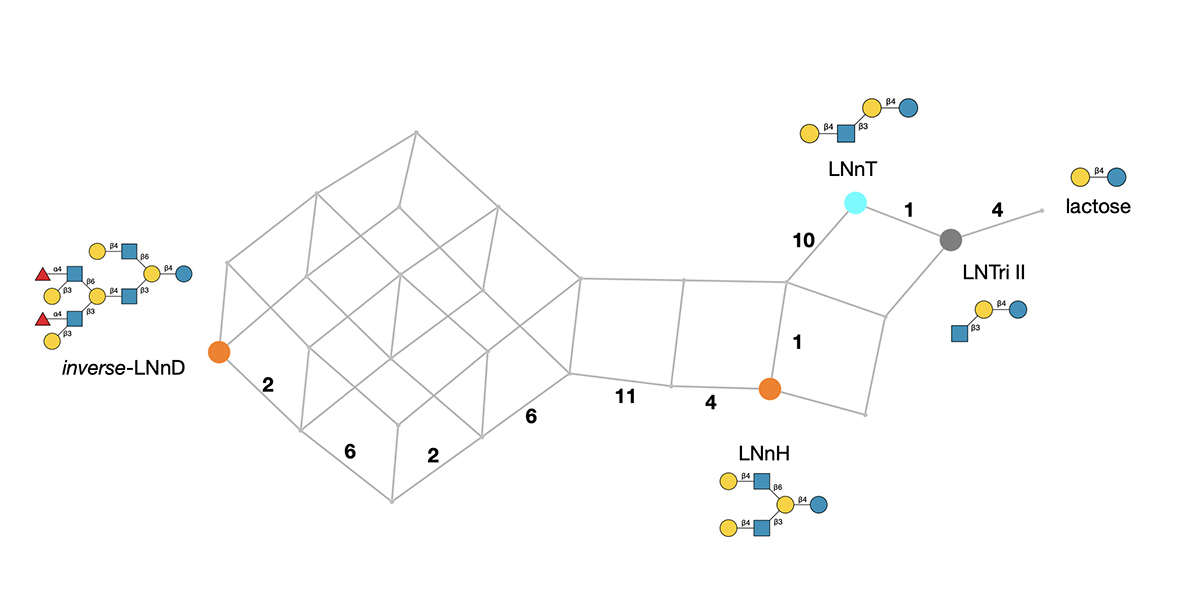
In the same way and with further iterations, any larger HMO can be traced from lactose, as shown in Figure 2, where a proposed biosynthetic network to the so-called inverse-LNnD (containing 12 monosaccharides) is detailed. Coloured nodes correspond to observed intermediates. Interestingly, very few of them are part of the original HMO dataset. Multiple routes to the same product are shown, with the enzyme numbers labelling the edges (reactions) of one possible route.
In the end, we showed that eleven enzyme activities of the model can account for 92% of all HMOs we analysed, that is, for which we have computed possible reaction networks. While the HMO-simulator can be tailored for two possible phenotypes, the single- and dual- knockouts of enzyme activities enabled us to rank the enzymes according to their degree of influence on the observed HMO population. The few HMOs that were not predicted by the model from a set of structures may point, either to novel enzyme activities unique to milk or otherwise to alternative functions of known enzymes.
Conclusion
As samples produced in various geographical, nutritional and health conditions accumulate, more HMOs will likely be identified in the future. In light of the first results of our analysis, we envisage that the HMO-enzyme simulator will be extended or adapted, as knowledge of the enzyme specificities improve. Our attempt to account for the largest collection of currently known HMOs in our model has raised several questions about individual and combined enzyme activities, some of which remain open. With its predictive power, the model can also be considered as a guide for the experimental synthesis of HMOs, which would potentially enable a relevant cocktail selection matching a variety of conditions. If tested, this could bring infant formula closer to reproducing the benefits of breast milk.
Details of this work are available here.
References
- Piecing Glycoscience together. Open Access Government. Issue 32
- Bioinformatics focus: Can we break the glycocode? Open Access Government. Issue 33
- https://www.ncbi.nlm.nih.gov/glycans/snfg.html
Please note: This is a commercial profile
© 2019. This work is licensed under CC-BY-NC-ND.











