Chair of Medical Cell BioPhysics Leon WMM Terstappen highlights why the interrogation of the cancer cells at the single cell level is required to truly understand cancer development
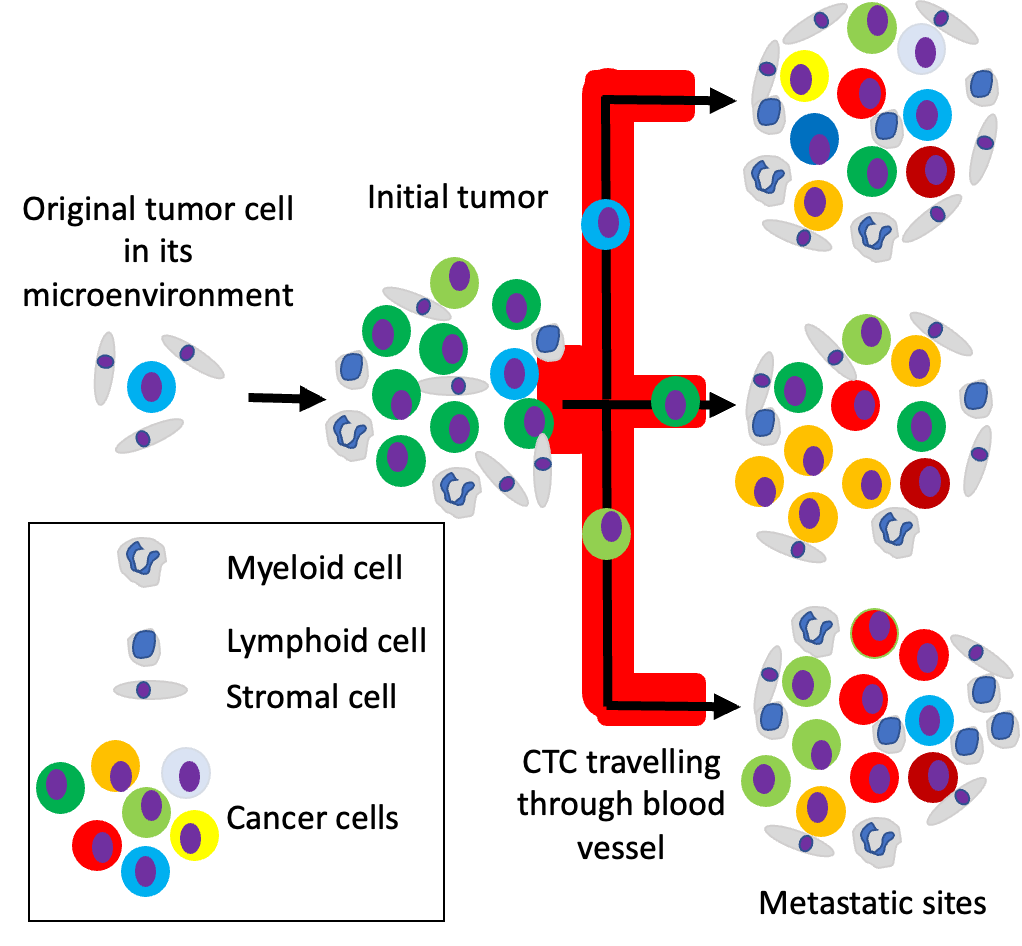
One of the main challenges for therapeutic success in cancer treatment is the heterogeneity of the cell composition in the tumour. When not all cancer cells are eliminated after treatment with curative intend the composition of the tumour, its load and distribution in the body will change over time as depicted in the figure.
Escape of some tumour cells from the initial tumour into the lymph and blood vessels and extravasation in other sites such as bone, liver, lung and brain stand at the core of cancer metastasis and is the lead cause of cancer death. At different metastatic sites, the cancer cells will adapt to the different local environments giving rise to cancer cell heterogeneity as depicted in the figure.
Identifying and selecting single cancer cells
Cell division of the already genetic altered cancer cells will further increase this heterogeneity and is being accelerated under the treatment the patient is receiving. Treatment options for cancer patients are rapidly increasing and a large proportion of novel drugs target specific cellular pathways.
A prerequisite for these drugs to work is that these pathways are active in the cancer cells of these patients and that the drugs can reach their target. Knowledge of the cellular composition of the cancer at the time systemic treatment is to be administered is thus essential to select the appropriate drug.
In the example shown in the Figure, a drug can for example “kill” a green tumour cell, inhibits the growth of an orange and red tumour cell but does not affect a blue or yellow cell. Giving this information a combination of therapies targeting all types of cancer cells is preferred. To obtain the cellular composition from all tumour sites is however not possible as taking biopsies from all sites is practically impossible.
One way to overcome this challenge is to gather the information from a blood sample as cancer cells and other tumour cell products from all these tumour sites travel through the blood. These so-called liquid biopsies hold great promise in making personalised medicine a reality.
A strong relationship between the load of cancer cells in the blood and the chance of survival of cancer patients with metastatic disease was demonstrated with the introduction of the CellSearch system and is recognised as one of the most important milestones in cancer this century.
Although monitoring Circulating Tumour Cells (CTC) load indicates whether or not the treatment is effective CTC enumeration does not provide the information which therapies might be effective in case the CTC load is not decreasing. To assess the presence of drug targets, the information can be extracted from these CTC.
The first step is to identify CTC among all the blood cells, which is quite a challenge. For example, when one CTC is present in 10ml of blood it will have to be identified and retrieved in the presence of approximately 5×107 leukocytes, 2×109 platelets and 5×1010 erythrocytes.
To overcome this challenge, enrichment techniques are applied making use of differences in physical, or immunological properties of the blood cells versus the CTC. The most successful techniques remove all erythrocytes and platelets and reduce the leukocytes with 3-5 logs. In the example used the detection technology needs to identify one CTC (when it’s not lost during the enrichment procedure) among 5×102 -104 leukocytes.
Immunofluorescence labelling is the most common technology used to differentiate between cells followed by fluorescent microscopic or flow cytometric detection. Antibodies recognising cytokeratin are frequently used to identify CTC and can be supplemented with antibodies against druggable targets such as the androgen, oestrogen, and her-2 receptor. The number of antibodies directed at different targets that can be detected simultaneously is however limited and much of the cancer cell heterogeneity is revealed after the individual cancer cells have been isolated.
At present, technology permits the determination of the DNA composition of the 3.2×109 nucleotides present in human cells at the single cell level whereas technology to determine and quantify the estimated 8-40 x104 proteins in a human cell does not exist yet.
At the core of single cell research is the ability to identify and select single cells. Several technologies are available or in development to extract single cells after immunomagnetic enrichment and fluorescently labelling. This includes flowcytometers with single cell sorting capability and fluorescence microscopes in combination with automated micropipettes, microchip-based digital sorting and self-seeding microsieves.
Each of these technologies has its pros and cons with none of them having a 100% yield of the original detected CTC which is problematic when few CTC are detected. Improving the yield of retrieving CTC that can be characterised is of great importance especially in those cases in which only few CTC are detected. Increasing the number of CTC available for analysis surely is benefitted using larger blood volumes, but the earlier in the disease the fewer CTC are present, making the challenge even greater.
Technological improvements advancing cancer cell research
For analysis of the DNA content of single CTC, the cells can be fixed making the sorting conditions needed to obtain single cells less stringent. To obtain functional information of the CTC including the sensitivity to potential drugs to be administered viable CTC are needed which not only increases the requirements for the isolation technology, but also lowers the number of available CTC as many are dead or are undergoing apoptosis.
To truly understand the development of cancer, measure the effects of treatment and adapt the treatment to either cure or turn it into a chronic disease while maintaining quality-of-life, interrogation of the cancer cells at the single cell level will be required.
Single cell research has provided us with new insights into the complexity of cancer formation and development. These insights are leading to new questions that can be answered but will need significant technology improvements to make single cancer cell isolation more affordable, more efficient and a more thorough analysis of their content in a timely fashion.
Please note: This is a commercial profile
© 2019. This work is licensed under CC-BY-NC-ND.