Emily Weinert, Assistant Professor of Chemistry at Emory University discusses an aspect of chemistry that concerns the growing body of work on the human microbiome
The varied nature of bacterial interactions with humans, playing essential roles in many aspects of human health, as evidenced by the growing body of work on the human microbiome,1 causing millions of cases of antibiotic-resistant bacterial infections each year,2 and the role of bacteria in crop and environmental health, has illuminated the need for an improved understanding of bacterial physiology. In particular, understanding the mechanisms by which bacteria sense their environment and alter cellular pathways to maximise survival has the potential to identify new chemical signals and macromolecular signalling cascades that can be targeted to modulate bacterial phenotypes, including growth and competition. However, our knowledge of the chemistry underlying many of these processes is still very minimal, despite the fact that understanding these pathways dictates our ability to rationally develop new methods to alter bacterial phenotypes.
To advance our knowledge of bacterial chemical sensing and signalling networks, extensive studies are needed to identify small molecules and proteins that bacteria use to respond to their environment. In addition, identifying other proteins involved in the signalling pathway and mechanisms of intercommunication between different sensing/signalling pathways is necessary to generate a holistic view of bacterial cellular function.
Without this fundamental knowledge, it is extremely challenging to predict how bacteria will respond to external stimuli, especially with regards to understanding what other pathways might interact, potentially muting or amplifying the phenotypic response. Generating this complex picture requires many types of experiments, from whole-cell methods that identify global effects, such as RNAseq and proteomics, to in vitro studies using purified components that allow for a molecular-level understanding of sensing mechanisms.
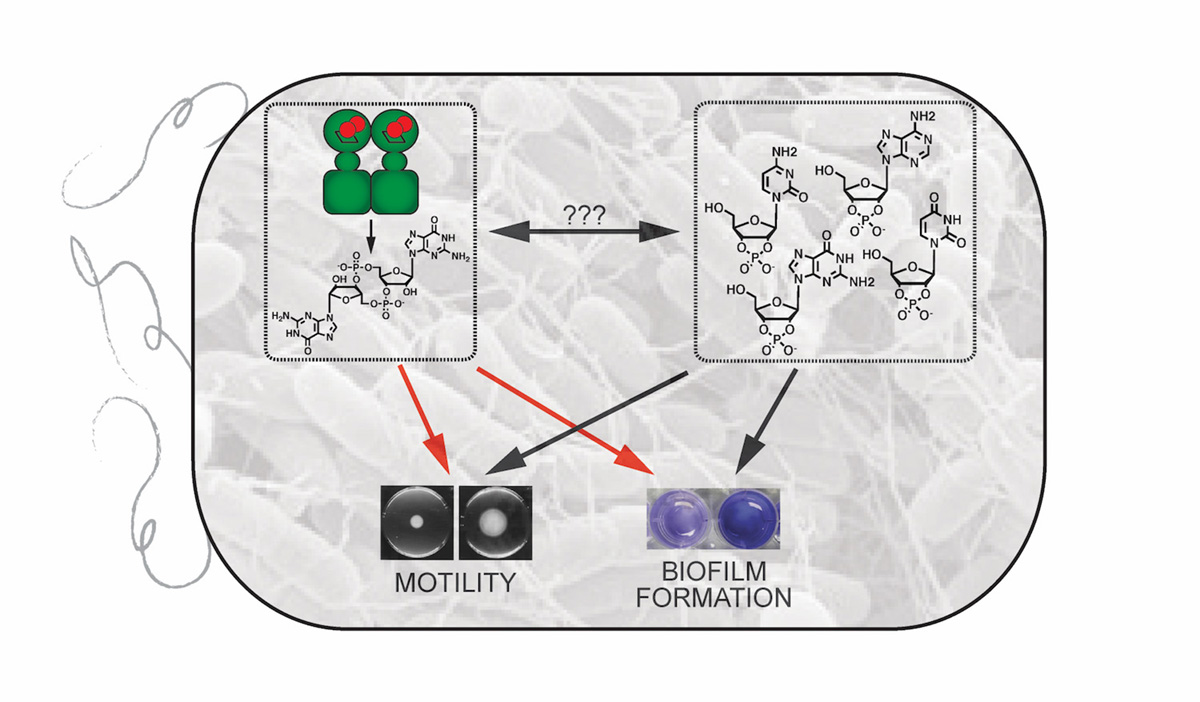
In my group, we are working to elucidate new bacterial signalling pathways using either a putative bacterial protein or small molecule as our starting point. To do so, my group has undertaken two interconnected projects involving previously understudied bacterial signalling pathways: 1) the mechanism and role of oxygen-sensing by diguanylate cyclase-containing globin coupled sensor (GCS) proteins and 2) the metabolism and cellular effects of 2’,3’-cyclic nucleotide monophosphates (2’,3’-cNMPs). By focusing on two different cyclic nucleotides found in bacteria, we can link results from our studies and provide insight into not only the individual pathways but the interlinked responses as well.
Towards this end, my group has focused on understanding signal transduction and downstream signalling for diguanylate cyclase-containing GCS proteins that serve as bacterial O2 sensors, since diguanylate cyclase produce cyclic dimeric guanosine monophosphate (c-di-GMP), a bacterial second messenger that controls biofilm formation.3 Working with a soft rot-causing plant pathogen, Pectobacterium carotovorum, we have demonstrated that the GCS protein controls O2-dependent motility, virulence factor production and the rotting of a plant host, highlighting the importance of GCS signalling for controlling key bacterial phenotypes.4 To understand how GCS proteins function, we have used a variety of biochemical, spectroscopic and microbiological techniques to shown that O2 binding causes conformation changes that alter GCS oligomerization and diguanylate cyclase activity.5
Furthermore, we have identified interfaces and residues involved in transmitting the O2 binding signal, which provides a starting point for developing tools to alter GCS activation. By providing key insights into signal transduction within GCS proteins, this work has improved our understanding of heme-based sensors and ligand-dependent signal transduction and has highlighted GCSs as potential new targets for antibacterial therapies.
Building on our interest in bacterial nucleotide signalling pathways, another project is focused on 2’,3’-cNMPs in bacteria and has allowed us to begin to identify proteins involved in 2’,3’-cNMP metabolism and downstream effects. The presence of 2’,3’-cNMPs in cell extracts was reported in the 1960s,6 but the results of those studies were discounted as artefacts of the extraction procedure shortly thereafter.
While quantifying 3’,5’-cNMP levels in various rat tissues, we discovered 2’,3’-cNMP isomers in rat organs, mammalian cells and Escherichia coli,7 providing an intriguing starting point to delve into the cellular chemistry and signalling of these nucleotides. By identifying the enzyme responsible for producing 2’,3’-cNMPs in E. coli and developing (bio)chemical tools to regulate 2’,3’-cNMP levels, we have demonstrated that these rediscovered cyclic nucleotides are generated during cytoplasmic RNA degradation and regulate bacterial biofilm formation and motility,8 suggesting potential interplay with c-di-GMP signalling pathways, which also regulate those phenotypes.
Our ongoing studies are focused on dissecting the cellular machinery related to 2’,3’-cNMPs, elucidating novel RNA decay pathways and highlighting novel stress-sensing pathways within prokaryotes, which may allow the 2’,3’-cNMP pathways to be engineered to control bacterial proliferation and virulence.
By identifying new molecules, proteins and pathways involved in bacterial signalling, the field can provide insights from the molecular to the organismal level, highlighting key bacterial phenotypes controlled by small molecules and the interplay between bacterial signalling pathways. Besides the potential identification of targets for the development of new anti-bacterial treatments, these types of studies can also provide insights into previously unexplored mechanisms of signal transduction and bacterial signalling pathways, improving our fundamental understanding of bacterial signalling.
References
1 Sommer, F., et al. Nat Rev. Microbiol. 2013, 11, 227.
2 Infectious Diseases Society of, A., et al. Clin. Infect. Dis. 2011, 52 Suppl 5, S397.
3 Walker, J. A., et al. Adv Microb Physiol 2017, 71, 133.
4 Burns, J. L., et al. ACS Chem. Biol. 2017, 12, 2070.
5 Burns, J. L., et al. Biochemistry 2016, 55, 6642.
6 Wade, H. Biochem. J 1961, 78, 457.
7 Jia, X., et al. Biomolecules 2014, 4, 1070.
8 Fontaine, B. M., et al. Biochem. J. 2018, 475, 1491.
Please note: this is a commercial profile
Emily Weinert
Assistant Professor of Chemistry
Emory University
Tel: +1 404 712 6865