Professor Ute Deichmann, Jacques Loeb Centre for the History and Philosophy of the Life Sciences, Ben-Gurion University of the Negev, discusses the history, current research and misconceptions about chromatin research and epigenetics
The correct expression of genes in the cell or in tissues at the right time is fundamental for the development and functioning of an organism. The basis for our molecular understanding of heredity was laid in the late 19th century when chromatin was first described by cytologists as the easily stainable threads in the cell nucleus. This became possible after dyes were available from the chemical industry, and the microscopy and biochemistry of the cell had made major progress.
Shortly after, biochemists defined chromatin as an association of a nucleic acid (later: DNA) and alkaline proteins (histones) into which eukaryotic genomes are packaged. This definition remains valid until today. For various reasons, biochemical research on DNA and chromatin proteins has stagnated since the early 20th century.
The new interest in the chemical basis of heredity in the 1930s, which led to the flourishing of molecular genetics, was based on research in bacteria and viruses that do not contain chromatin, because their chromosomes or DNA, respectively, do not include complexes with proteins.
Only when, in the 1960s, molecular biologists began to explore more complex systems, such as eukaryotic cells and development, was research into the structural analysis and function of chromatin resumed with a focus on histone modifications. Chromatin is now known to play an important role in the highly complex machinery of gene regulation in eukaryotes that includes several protein complexes and long non-coding RNA.
The term epigenetics that was introduced in 1942 for the causal interactions between genes and their products during development, has drastically changed its meaning over time. It is now used for a wide range of research related to the complex machinery of gene regulation that includes DNA and chromatin modifications by methyl-, acetyl and other groups.
Histone modifications
The pioneers of modern chromatin research were Vincent Allfrey and Alfred Mirsky, who in the 1960s, confirmed histones’ inhibitory effect on transcription and showed that their modifications by acetylation and methylation apparently alleviated this effect. However, they could not show whether this was a causal relationship or an accidental correlation.
In the 1990s, new research on chromatin structure modification established a connection between chromatin structure and its function. It was shown in yeast that a mutation of enzymes responsible for histone acetylation had a direct effect on yeast growth. Enzymes that move the nucleosomes around on DNA, chromatin remodeling enzymes, which had significant phenotypes, were also detected through yeast genetics.1
Histone modifications consist of small molecules, mostly methyl or acetyl groups, bound to certain residues of histone tails. The responsible enzymes are not DNA sequence-specific, i.e., they need DNA-specific factors to recognize the histones in question. Histone modifications are sometimes transmitted by cell division and, in rare cases, also by the germ line. However, the modifications are not stable and not faithfully copied, and they disappear after a few cell generations.
As of now, there is no evidence that these modifications affect gene activity, but there is ample evidence that histone acetylation is generated as a consequence of transcription: The binding of transcription factors precedes histone acetylation that then reduces the affinity of the histone proteins for DNA, thus rendering transcription events easier (figure 1).1
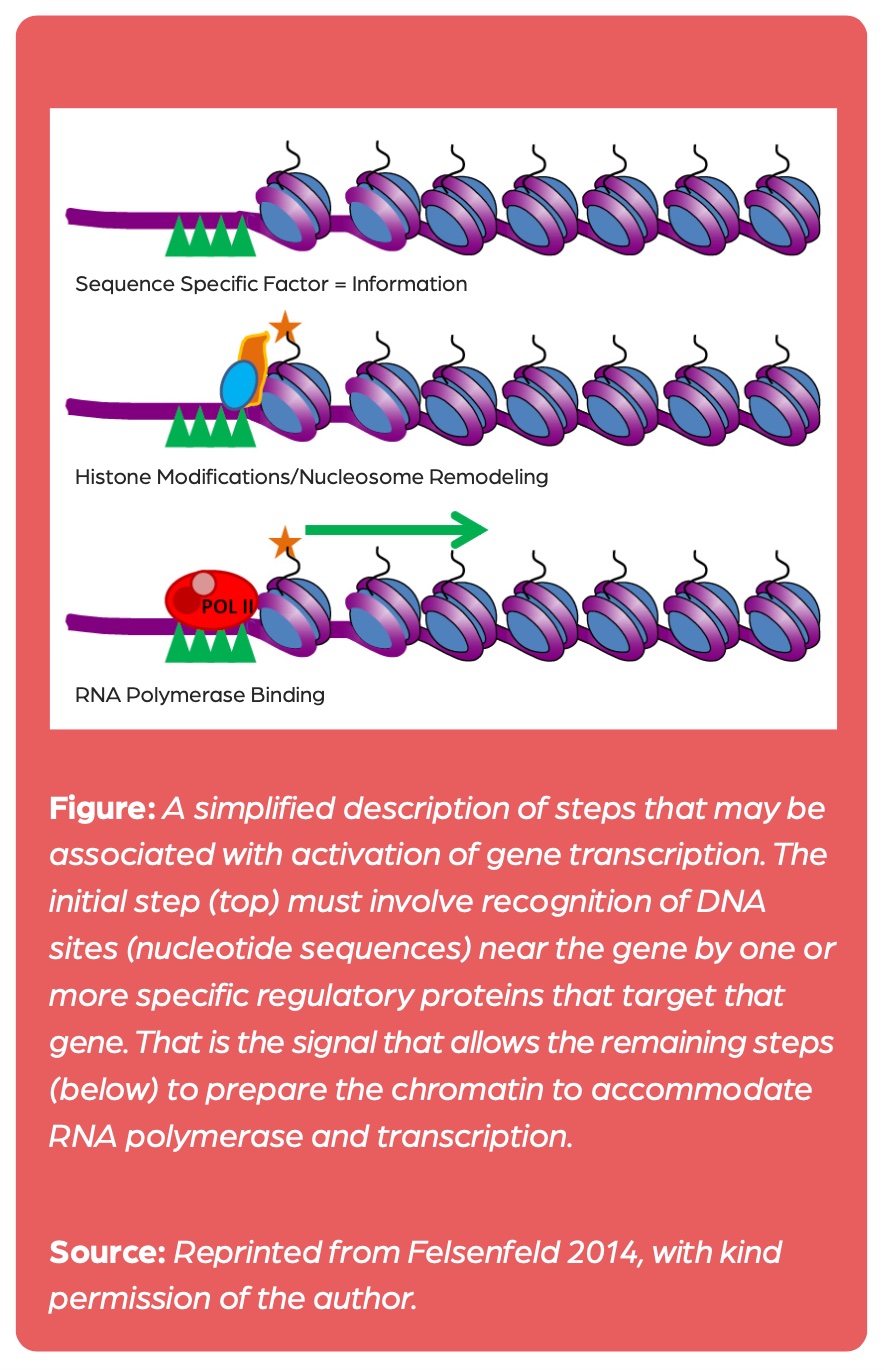
Since the late 20th century, research on chromatin modifications has also been conducted under the label epigenetics.
DNA Methylation
The discovery of imprinted genes (in mice and men) in the 1990s led to a close association of DNA methylation with epigenetics – DNA methylation had been studied without connection to epigenetics since the 1970s.
But research labelled “epigenetics” remained marginal until heritability was added to its definition. Robin Holliday proposed that patterns of DNA methylation were heritable through cell division and that there were also examples of epigenetic inheritance through the germ line. Therefore (de-) methylation of DNA should be called epigenetic.
However, the idea of transgenerational epigenetic inheritance in higher organisms is highly problematic. Moreover, most recent definitions of epigenetics do not distinguish between the propagation of modifications through cell division (thus helping to maintain a pattern of gene expression) and other cases in which the modifications are simply part of the transcriptional apparatus that is its biochemistry.1
It is generally accepted that DNA methylation is necessary for the suppression of transposons, i.e., parasitic DNA elements that can jump to different places in the genome, imprinting, and X chromosome inactivation. But the role of DNA methylation and histone modification in the biochemical events that regulate genes is still not clearly established. There are groups of organisms such as nematodes (small roundworms) and certain insects, such as Drosophila, which do not methylate their genomes.
It is undisputed that DNA methylation does not silence active promoters of genes (DNA regions where transcription is initiated) but affects genes that are already silent. It is also known that eukaryotic cells use many mechanisms to shut down and maintain repression of gene activity. According to Timothy Bestor et al., the available data do not support “the existence of a biochemical system that regulates embryogenesis by programmed methylation and demethylation of regulatory sequences.”2
The current usage of the term epigenetics
Research labelled epigenetics has experienced a rapid rise in the 21st century, accompanied by an increasing diversity in researchers’ understanding and definitions of epigenetics.
It now includes research on histone and DNA-modifying enzymes, nucleosome remodelers, histone chaperones, chromatin-binding proteins to facilitate transcription factor and polymerase action, and the role of long non-coding RNA and small interfering RNA in transcriptional regulation.
Geneticist and epigeneticist John Greally urges researchers to clarify the way in which they use the term epigenetics, and he also made it clear what should not be labelled epigenetics: “If we mean epi + genetics—the layer of information beyond the genome—it is unclear why we don’t just say ‘transcription regulation’ instead.
If our definition is basically a proxy for the mediation of environmental responses, this should not be equated with epigenetic processes at all.”3
Common misconceptions and unsupported speculations and epigenetics
- Epigenetics does not relativize the importance of genes
- As was shown above, it is impossible to separate epigenetics from genetics, i.e., DNA sequence- specific events: The enzymes that attach modifying molecules to DNA or histones do not recognize specific DNA sequences.
- Therefore, specific regulatory factors such as transcription factors or proteins from the polycomb group are necessary to target the transcriptional regulatory machinery to specific places on the DNA. Development, for example, is based on the specific turning on and off of sets of genes over time. Transcription factors also mediate environmental influences on gene activity and maintain cellular memory in a sequence-specific way.
- The environment has no lasting impact on the change of epigenetic marks
- Epigenetics has often been used to support claims of a direct impact of environmental factors on hereditary characteristics. The environment can act on the phenotype through transcriptional regulation and cellular differentiation and these processes can involve epigenetic factors.
- Much of the resulting stability and cellular memory is based on gene regulatory networks involving feedback loops. But chromatin modifications are insusceptible to the direct influence of environmental factors, apart from some synthetic inhibitors of chromatin modifying enzymes. Rather, environmental factors affect gene expression through signalling cascades, which activate or repress transcription factors.4
- Despite many misconceptions about the mechanisms involved, epigenetic defects contribute to human disease. But most epimutations, i.e., aberrant chromatin states without DNA sequence changes, are not the cause of the disease but part of the mechanism by which a genetic mutation causes disease.4
- An example is autism spectrum disorders like Rett and Fragile X syndromes that have been shown by Adrian Bird to be caused by mutations.5,6 The Rett syndrome is caused by mutations in the gene that encodes the methyl CpG binding protein 2 (MECP2) that reads DNA methylation and plays an important role in nerve cells. If it is mutated, the DNA methylation doesn’t change, but the protein that reads it is lost.
- Transgenerational epigenetic inheritance is questionable in humans
- Many of the suggested examples of epigenetic inheritance in humans concern inter- rather than transgenerational effects and rarely exclude DNA sequence changes as the underlying cause for heritability. Parental or intergenerational effects occur when the uterus is exposed to toxins, viruses, detrimental nutritional, or hormonal environments that directly affect the developing embryo and its germline.
- This exposure usually impacts the first generation, but occasionally also grandchildren. In contrast, transgenerational effects relate to generations that were not exposed to the initial environmental trigger, i.e., to great-grandchildren and beyond. Intergenerational effects occur in humans and other mammals, but there are two rounds of efficient reprogramming and erasure of DNA methylation in the development of totipotent cells in the early embryo as well as during germ cell differentiation.
- It is widely believed that this reprogramming prevents the inheritance of most of the epigenetic marks. According to Edith Heard and Robert Martienssen “although much attention has been drawn to the potential implications of transgenerational inheritance for human health, so far there is little support.”7
- The maternal environment can have long-lasting effects on our health. In the Dutch hunger winter, for example, severe undernourishment affected pregnant women, their unborn offspring, and the offspring’s fetal germ cells. But the increased incidence of cardiovascular and metabolic disease observed in the first generation is not due to the transmission of epigenetic information through the maternal germline, but a direct consequence of the exposure in the uterus, a phenomenon called “fetal programming” or – if fetal germ cells and the second generation are affected – “intergenerational inheritance.”8
In summary, epigenetics cannot be clearly separated from genetics because epigenetic mechanisms such as DNA methylation are dependent on DNA sequence-specific events. Gene activation and repression during development are controlled not by DNA methylation but by well-established protein- and RNA- based mechanisms.
Therefore, research on chromatin modification, DNA methylation, etc. did not replace genetic and genomic research. Chromatin research and epigenetics, however defined, are good examples of how research directions and medical applications are expanded, and how new strands of research are synthesized with established fields – genetics and cell biology as well as the well-known principles of gene regulation.
References
- Felsenfeld, Gary. 2014. The evolution of epigenetics, Perspect Biol Med 57(1): 132–48.
- Bestor TH., Edwards, JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development, PNAS. 2015;112: 6796–9.
- Greally JM. A user’s guide to the ambiguous word ‘epigenetics’, Nature. 2018;19: 207–8.
- Horsthemke B. A critical appraisal of clinical epigenetics, Clinical Epigenetics. 2022;14: 95. https://doi.org/10.1186/s13148- 022-01315-66 Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms, Cell. 2014;157: 95–109.
- Bird A. Interview by U Deichmann May 28, 2018. https://in.bgu.ac.il/en/loeb/OHP/Pages/Adrian-Bird.aspx.
- Bird A. Proteins that interpret genomic signals to stabilise cell identity. Paper at the Workshop “Plasticity and Constancy in Development and Evolution,” Ben-Gurion University of the Negev, May 9-10, 2022.
- Heard E. Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms, Cell. 2014;157: 95–109.
- Horsthemke B. A critical view on transgenerational epigenetic inheritance in humans, Nature Communications. 2018;9: 2973.
To read and download the full eBook “Chromatin research and epigenetics: History, Current Research, and Misconceptions”, click here


