Prof Thomas Jaki from the Medical and Pharmaceutical Statistics Research Unit at Lancaster University sheds light on dose-finding trials
The primary aim of dose-escalation trials (Phase I trials) is often to find the largest dose of a treatment that can be administered safely, often called the maximum tolerated dose (MTD). Phase I studies are crucial in the development of a novel treatment as the agent is often being given to humans for the first time. To ensure the safety of the patients in the study, they tend to start at doses much lower than the anticipated highest safe dose and only slowly increase the dose until the target dose is found. Dose-escalation studies have been described by Edler (2) as “first-into-human studies where safety is paramount and close monitoring essential”.
Algorithmic designs
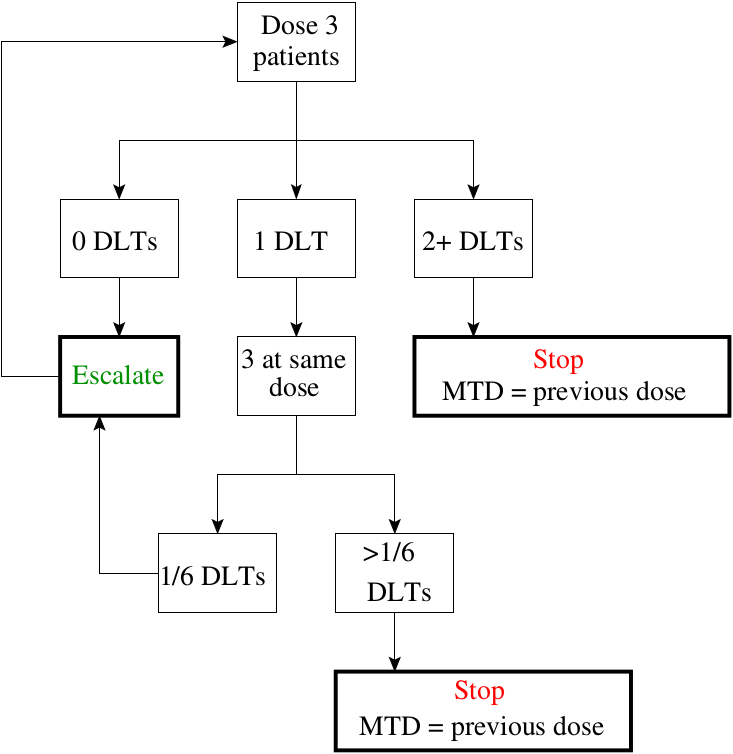
Rule-based designs, such as the 3+3 design (1) (illustrated in Figure 1), continue to be popular even though they can perform extremely poorly. The alternative to these rule-based designs are (Bayesian) model based approaches which use a statistical model to describe the dose-response relationship (3). Here we list the main arguments why 3+3 and similar rule-based A+B designs should not be used for phase I dose escalation studies.
- A+B designs lead to more patients being treated at sub-optimal doses (i.e. patients either receive a sub-therapeutic or an overly toxic dose);
- There is only a one in three chances to obtain the correct dose at the end of a 3+3 design, compared to better than 1 in 2 for model-based designs;
- In contrast to model-based designs, A+B designs do not offer a measure of variability in the final recommendation and hence do not provide any measure of confidence in the recommendation;
- A+B designs are highly inflexible and therefore need ad-hoc changes to cope with many situations observed in practice (e.g. drop-out of patients);
- A+B designs use only the information from the last cohort of patients whereas model-based designs make use of all data accumulated.
1 Carter SK. Study design principles for the clinical evaluation of new drugs as developed by the chemotherapy programme of the National Cancer Institute. In: Staquet MJ. The design of clinical trials in cancer therapy. Mount Kisco, NY: Futura Publishing Co, 1973:242-89.
2 Edler L. Overview of phase I trials. In: Crowley J. Statistics in clinical oncology. New York: Dekker, 2001:1-34.
3 Cheung YK (2011) Dose Finding by the Continual Reassessment Method. Chapman & Hall/CRC, Boca Raton, FL. ISBN 9781420091519.

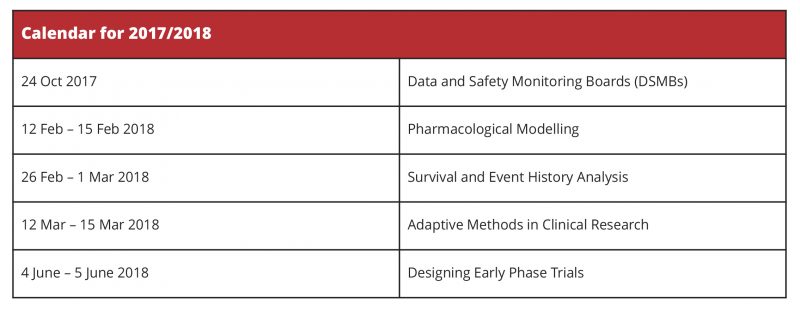
For details of these and other courses offered by the Postgraduate Statistics Centre, go to http://www.lancaster.ac.uk/maths/postgraduate/short-courses/ or contact Angela Mercer, Postgraduate Statistics Centre, Department of Mathematics and Statistics, Fylde College, Lancaster University, Lancaster LA1 4YF UK.
All these professional development courses, as well as many other, may be commissioned by a company or organisation, tailored to meet your special requirements and presented at a location of your choice. Please contact us to discuss your specific requirements or for further information.
Please note: this is a commercial profile
Prof Thomas Jaki
Professor of Statistics
Medical and Pharmaceutical Statistics Research Unit, Lancaster University
Tel: +44 (0)1524 59 23 18