Here we explore the massive impact nucleic acid chemistry has had on the manufacture of synthetic DNA and RNA sequences whilst explaining the chemistry behind the chemical synthesis of DNA
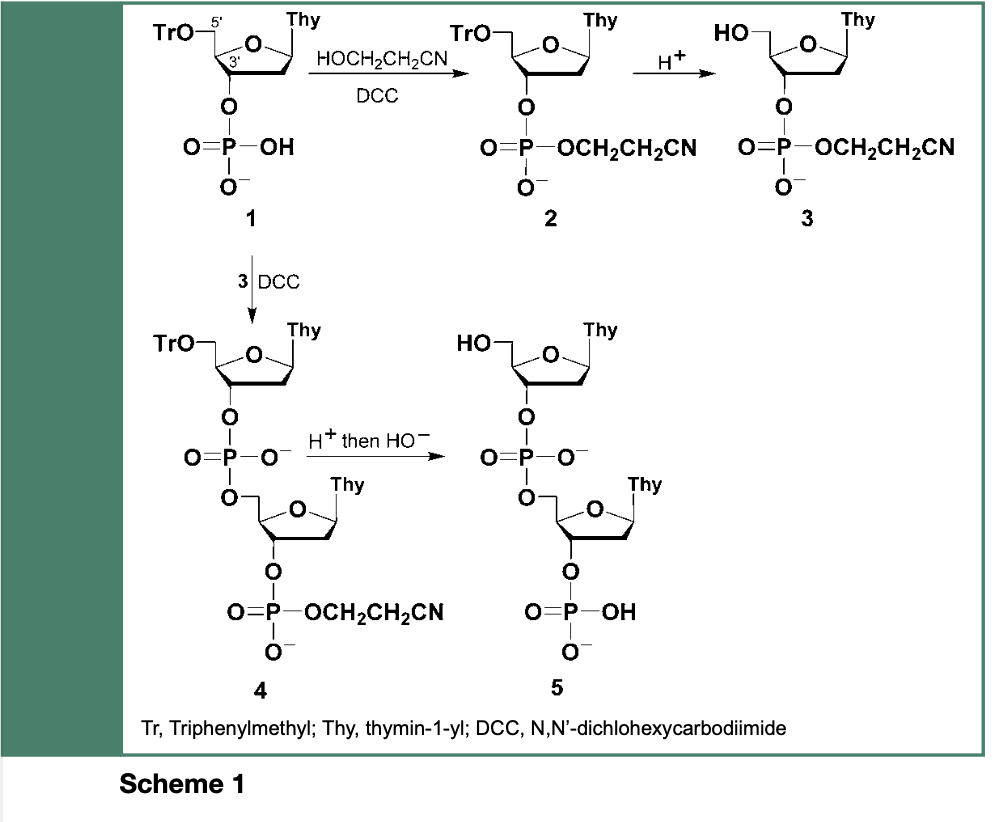
Nucleic acid chemistry has a long history. Indeed, Todd and coworkers initiated, in the early 1950s, an extensive study of nucleoside polyphosphates (e.g., ADP, ATP), which later led Gilham and Khorana to implement the use of carbodiimides or arenesulfonyl halides to facilitate esterification of P(V) oxidation level deoxyribonucleoside phosphomonoesters to their corresponding phosphodiester derivatives 2 and 3 (Scheme 1).
The phosphodiester approach to the chemical synthesis of polydeoxyribonucleotides
These deoxyribonucleoside phosphodiesters allowed Khorana and coworkers to synthesize, in the early 1960s, sequence-specific oligonucleotides upon activation of 1 with N,N’-Dicylohexexylcarbodiimide (DCC) and its esterification upon reaction with the hydroxyl function of 3 to provide the dinucleotides TpTp 4 and 5 as illustrated in Scheme 1. When employing the phosphodiester chemistry with 5’-O-protected and appropriately amine protected deoxyribonucleosides (i.e., cytidine, adenosine or guanosine), the elaborate and challenging chemical synthesis of polydeoxyribonucleotides blocks was conducted.
Enzymatic conversion of these blocks to polyribonucleotides led to in vitro production of all the proteinic codons needed to elucidate the genetic code. Complete unraveling of the genetic code was achieved by Khorana and his colleagues in 1966. While employing the phosphodiester chemistry, Khorana completed the total synthesis of a gene in 1972 and demonstrated that this artificial gene was functional in a bacterium. Although the phosphodiester chemistry had contributed to the total synthesis of tRNA genes, the purification of negatively charged gene products was tedious and time- consuming. A synthetic method was sought for the chemical synthesis of DNA sequences that will allow production of uncharged DNA or RNA sequences to facilitate the purification of these nucleic acid sequences.
The phosphotriester approach to the chemical synthesis of polydeoxyribonucleotides
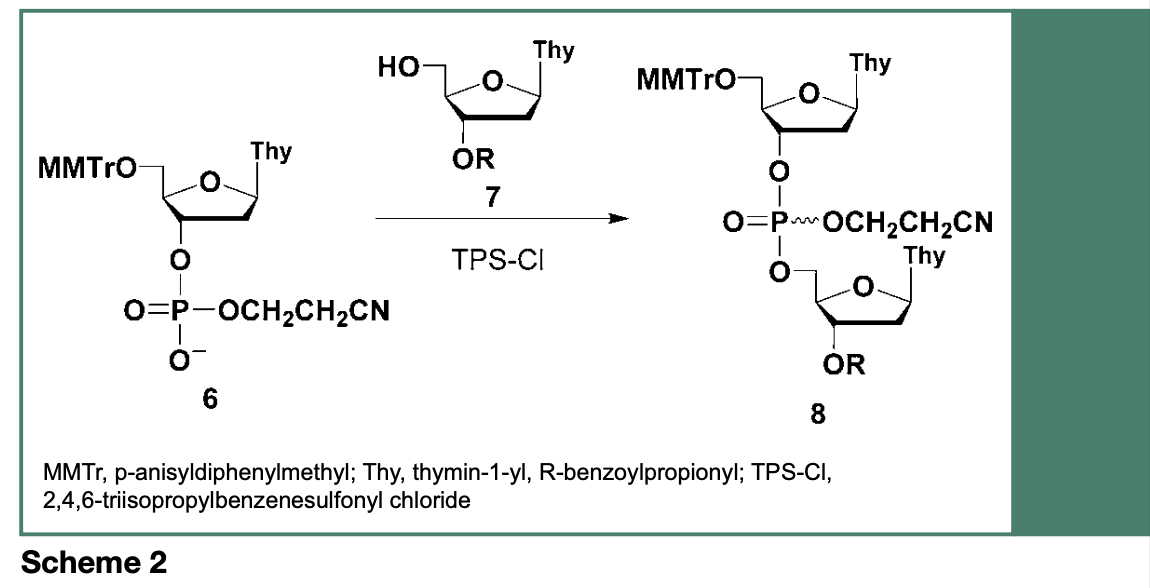
In the late 1960s, Letsinger and Ogilvie reported the reaction of a deoxyribonucleoside phosphodiester (6) with a 3’-O-protected deoxyribonucleoside (7), in the presence of 2,4,6-triisopropylbenzenesulfonyl chloride (TPS-Cl), yielding an uncharged dinucleoside phosphate triester 8 (Scheme 2), which was easily separated from the negatively charged 6 and isolated by silica gel chromatography. This synthetic process accurately represents the tenet of the phosphotriester approach to the chemical synthesis of DNA sequences, which led to the implementation of a strategy for the stepwise synthesis of mixed polydeoxyribonucleotides.
The chemical stability of the 2-cyanoethyl phosphate protecting group became questionable, particularly in the context of large polydeoxyribonucleotide syntheses. The search for chemically stable phosphate protecting groups and for methods to be used for their eventual removal, had been intensely sought. Aryl groups have been proposed by Reese and others in the late 1970s and early 1980s; the 2-chlorophenyl group was demonstrated to be the preferred aryl phosphate protecting group for oligodeoxyribonucleotide synthesis by the phosphotriester method.
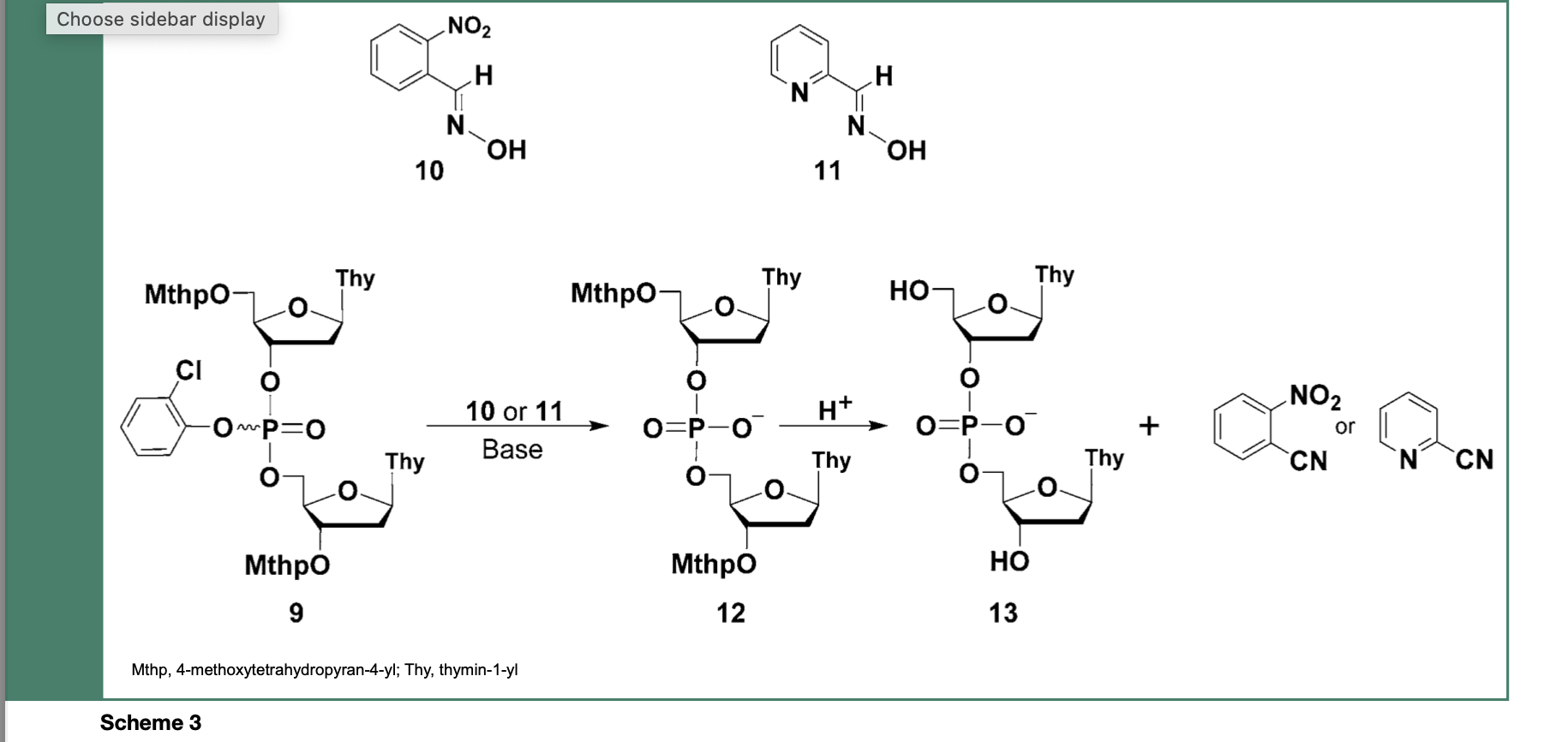
Technically, the 2-chlorophenyl phosphate protecting group was completely stable during chain assembly but readily cleaved, within 30 min, from the dinucleoside phosphate triester 9, upon treatment with E-2-nitrobenzaldoxime (10) or syn-pyridine-2-carboxaldoxime (11) salts to produce the dinucleoside phosphate diesters 12 and 13 along with 2-nitrobenzoni- trile or 2-cyanopyridine as a deprotection side- product (Scheme 3).
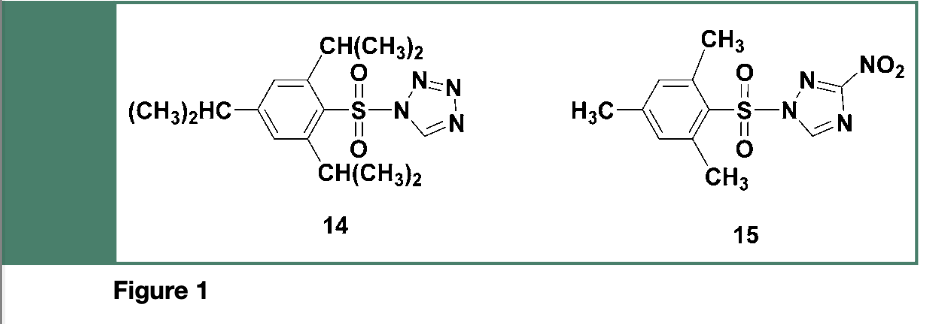
Given that the crucial step to be performed when using the phosphotriester method, is the activation of deoxyribonucleoside phosphodiesters (e.g., 6) by arylsulfonyl chlorides (e.g., TPS-Cl), sulfonylation of the 5’-hydroxyl of incoming nucleosides (e.g.,7) or nucleotides is to be avoided. For this reason, sterically hindered arylsulfonyl azolides, such as the 2,4,6 triisopropylbenzenesulfonyl tetrazolide 14 or the more stable 2,4,6-trimethylbenzesulfonyl 3-nitro 1,2,4- triazole 15 (Figure 1), both had demonstrated to be very effective as coupling reagents for phosphate triester syntheses, due to their lesser reactivity toward hydroxyl functions.
It has also been reported that the presence of catalysts, such as 1-methylimidazole or 4-dimethyaminopyridine-1-oxide, significantly improved the kinetics of coupling reactions;
a complete coupling reaction was achieved within 2 min when arysulfonyl chlorides were employed for activation of deoxyribonucleoside phosphodiesters toward the synthesis of dinucleoside phosphate triesters.
Such research achievements had undeniably fueled the development of innovative technologies for the chemical synthesis of nucleic acid sequences, as defined below.
The solid-phase strategy for the chemical synthesis of DNA and RNA sequences
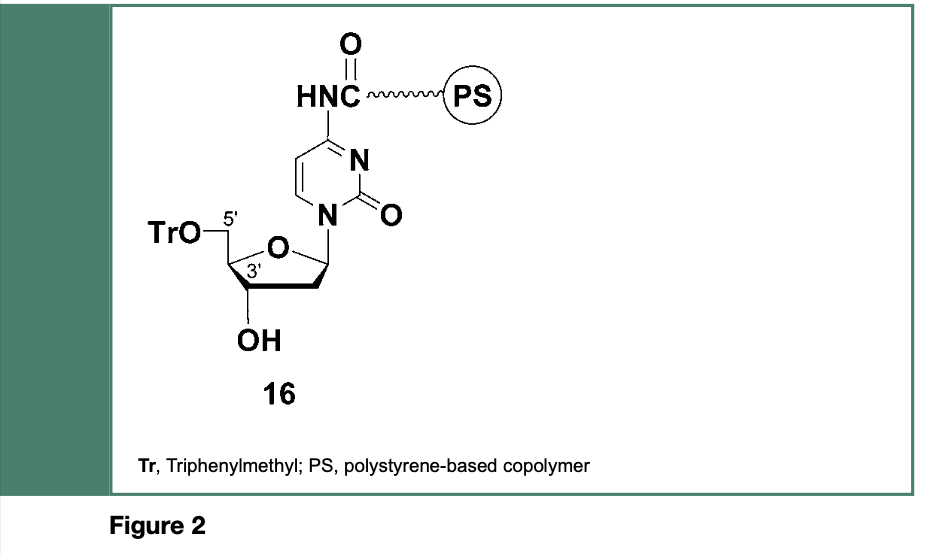
It is important to mention that the groundbreaking strategy, linked to the development of the phosphotriester approach to the chemical synthesis of oligodeoxyribonucleotides, (Letsinger & Ogilvie, 1969) was originally employed for the stepwise synthesis of di-, tri- and tetranucleotides on a polymeric support composed of styrene, p-vinylbenzoic acid and p-divinylbenzene (Letsinger & Mahadevan 1965, 1966); exposure of the chlorocarbonyl functions of the copolymer to the exocyclic amine, 3’-hydroxyl or 5’-hydroxyl functions of adequately protected nucleosides led to, for example, the copolymer-bound deoxyribonucleoside 16 (Figure 2).
The phosphodiester or the phosphotriester method has been employed for polymer- supported stepwise oligodeoxyribonucleotide (DNA) synthesis. The full-length DNA sequence along with the shorter-than-full-length DNA sequences produced from these methods, are cleaved from the solid support 16 under basic conditions.
The main advantage of the stepwise polymer-supported DNA synthesis is that the DNA chains can be freed from solvents and excess reagents by filtration after each chain extension step. The novelty of polymer-supported DNA synthesis attracted considerable interest and research efforts, which led to the development of a plethora of organic supports including, in addition to polystyrenes, polyamides and polysaccharides. However, organic supports had the propensity to swell, when exposed to organic solvents, thereby restricting the diffusion of reagents and solvents through these supports; such shortcomings prohibited the use of organic supports for automated solid-phase synthesis of oligodeoxyribonucleotides and shifted research interests toward the use of inorganic solid supports for automated solid-phase DNA synthesis.
Indeed, the silica-based material used for high-performance liquid chromatography (HPLC) is porous and does not swell when in contact with organic solvents, thus allowing reagents and solvents to freely diffuse into and out of the support. The use of HPLC grade silica-based supports (Köster, 1972, Matteucci & Caruthers, 1980, 1981) led to consistently high coupling yields and was deemed acceptable for automated solid-phase synthesis of DNA sequences. Nonetheless, amorphous silica-based support was soon replaced with a high-silica glass having pores with a specific size distribution (i.e., controlled pore glass or CPG); this type of support has been and is still currently being extensively used in the automated synthesis of oligodeoxyribonucleotides and oligoribonucleotides.
Although the phosphotriester approach was attractive for the synthesis of DNA sequences in solution, the method was observed to be somewhat sluggish and inefficient. By the mid- 1970s, Letsinger and coworkers developed and implemented an innovative P(III) oxidation level approach to the synthesis of DNA sequences. The method consisted of phosphitylating a 5’-O-protected deoxyribonucleoside (17) with a phosphorodichloridite (18) to provide the corresponding deoxyribonucleoside- 3’-O phosphorochloridite intermediate 19, which was rapidly (~ 5 min) formed at -78 °C (Scheme 4) to provide the symmetrical (3’→3’) dinucleoside phosphite triester 20.
Addition of 3’-O-protected deoxythymidine (21), followed by an aqueous solution of iodine in the presence of 2,6-lutidine, led to the formation of the desired (3’→5’) dinucleoside phosphate triester 22 along with the symmetrical (3’→3’) and (5’→5’) dinucleoside phosphate triesters 23 and 24 (due to unreacted 18). The preparation of deoxyribonucleoside chlorophosphites (e.g., 19) from reactive bifunctional phosphitylating reagents (e.g., 18) had to be performed at low temperature in the absence of moisture under an inert atmosphere to prevent hydrolysis an air oxidation of 19.
Furthermore, the concomitant formation of symmetrical (3’→3’) and (5’→5’) dinucleoside phosphate triesters when using deoxyribonucleoside chlorophosphites was undesirable and deterred the routine use of such reagents in the automated solid-phase synthesis of oligodeoxyribonucleotides. These problems were resolved in the early 1980s through the development and implementation of deoxyribonucleoside phosphoramidites, as an innovative new class of reagents for the chemical synthesis of DNA (Beaucage & Caruthers, 1981) and subsequently RNA sequences.
The phosphoramidite approach to the automated solid-phase synthesis of deoxyribooligonucleotides
Unlike deoxyribonucleoside chlorophosphites, deoxyribonucleoside phosphoramidites
are stable to hydrolysis and air oxidation. This unique class of reagents have been originally prepared from the reaction of 5’-O-protected thymidine (17) with chloro-(N,N- dimethylaminomethoxyphosphine and N,N- diisopropylethylamine (Scheme 5) to yield the deoxyribonucleoside phosphoramidite 25, near quantitatively, as a stable solid product.
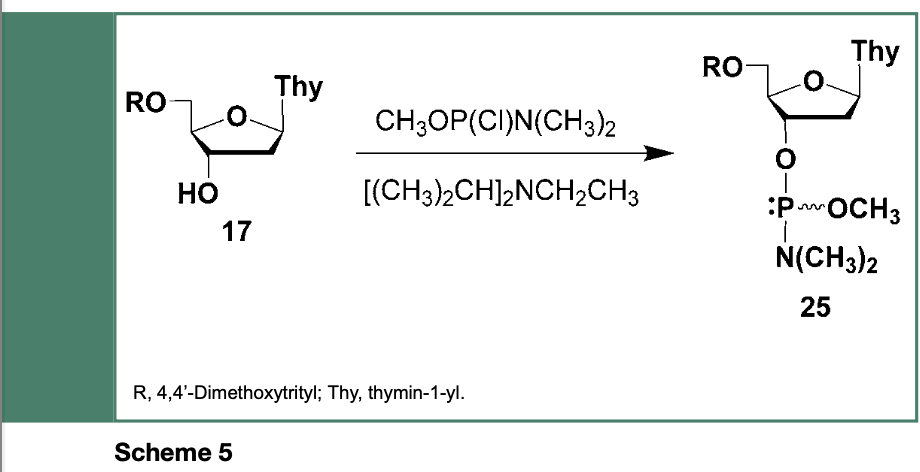
However, the use of the deoxyribonucleoside phosphoramidite 25 in the automated synthesis of DNA sequences was not optimal because 25 and its congenerics were not totally stable in acetonitrile due to the presence of acidic contaminants, which were difficult to completely remove while employing chromatography techniques.
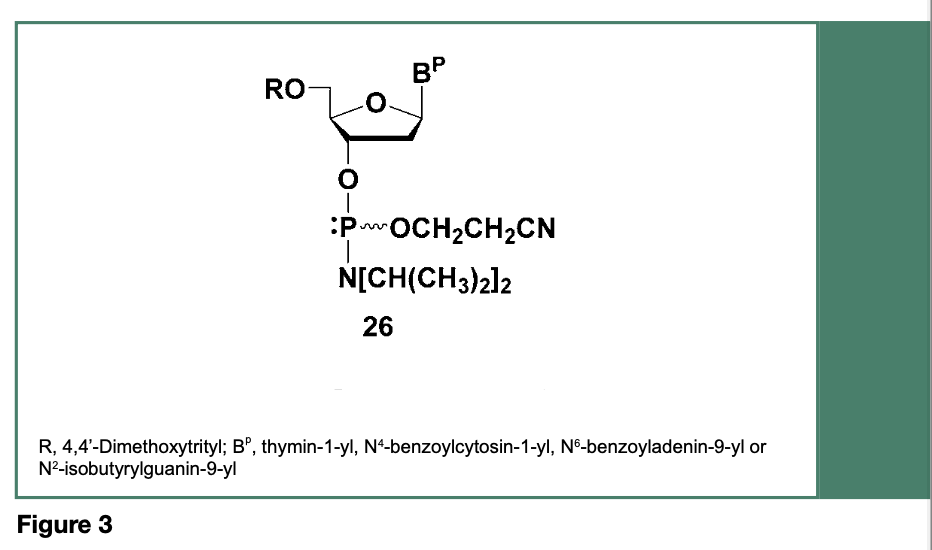
This shortcoming prompted research efforts toward the development and implementation of deoxyribonucleoside phosphoramidite monomers with improved P(III) O-alkyl and N,N-dialkylamino functions to provide enhanced stability, often at the expense of phosphoramidite reactivity. Thus, replacement of the original N,N-dimethylamino function with other N,N-dialkylamino or N,N-cycloalkylamino groups and replacement of the methoxy P(III) protection with a variety of functionalized alkoxy groups (Beaucage & Iyer, 1992) led to, as illustrated in Figure 3, the most popular and currently commercially available deoxyribonucleoside phosphoramidites (26), which have been used since the mid-1980s.
When activated with a weak acid, (e.g., 1H-tetrazole), the phosphoramidites 26 have been used for the synthesis of a relatively large (150-mer) DNA sequences on non-porous silica microbeads (Seliger et al., 1989) with stepwise coupling yields averaging 98-99%. Since the implementation of the deoxyribonucleoside phosphoramidites 26 more than 40 years ago, the use of these phosphoramidites for the manufacture of DNA sequences is still going strong.
The phosphoramidite approach to the automated solid-phase synthesis of ribooligonucleotides
Given that a protecting group is required for the 2’-hydroxyl of ribonucleosides, the chemical synthesis of ribooligonucleotides is de facto more complex than that of deoxyribooligonucleotides; the selected 2’-O-protecting group must remain stable during assembly of the RNA sequence and, for most synthetic strategies, be quantitatively cleaved when needed, under conditions that will not result in the cleavage or migration of internucleotide linkages. The search, for a 2’-O-protecting groups that might be stable under the acidic conditions required for complete removal of the 5’-O-pixyl or 5’-O-dimethoxytrityl groups, identified, as shown in Figure 4, the tert-butyldimethylsilyl (TBDMS), 1-(4-chlorophenyl)-4-ethoxypiperidin- 4-yl (Cpep) and 1-(2-fluorophenyl- 4-methoxypiperidin-4-yl (Fpmp) as 2’-O-protecting groups for ribonucleosides (Beaucage & Reese, 2009).
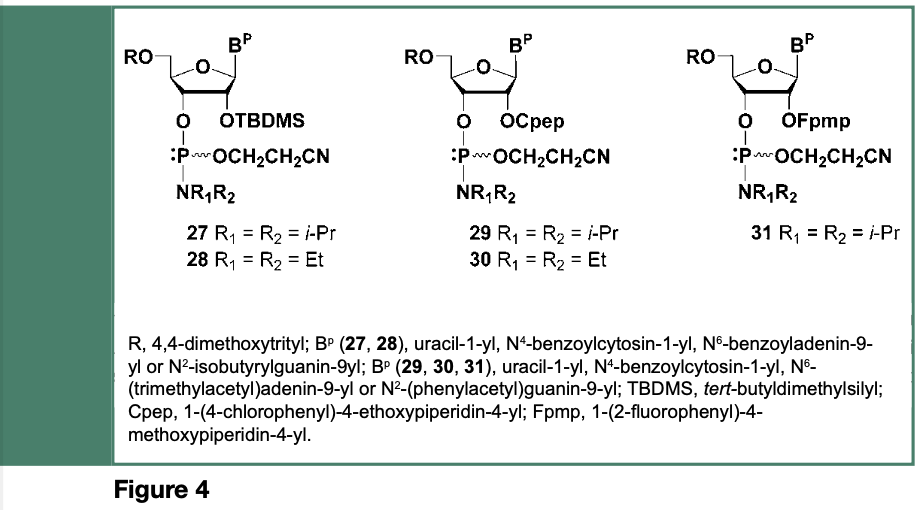
The above 2’-O-protected ribonucleoside phosphoramidites were successfully employed in the solid-phase synthesis of oligoribonucleotides with average stepwise yields of 98% leading to chain lengths exceeding far more than 20-mers; an RNA sequence (77-mer) corresponding to an E. coli tRNAfMet analogue was reported by Ogilvie et al., in 1988.
The H-Phosphonate approach to the automated solid-phase synthesis of deoxyribo- and ribo-oligonucleotides
Although the synthesis of tri- and tetranucleotides was achieved, in the early 1970s, using nucleoside H-phosphonates covalently bound to polystyrene-based polymer (Kabachnik et al., 1971), the yields of these polydeoxyribonucleotides were low. The use of H-phosphonate intermediates in the solid-phase synthesis of oligodeoxyribo-nucleotides was reexamined in the mid-1980s. An improved synthesis of deoxyribonu-cleoside H-phosphonates was achieved from the reaction of 5’-O- and exocyclic amine protected deoxyribonucleosides (32) with tri(1H-1,2,4-triazol-1-yl)phosphane (33) to provide after hydrolysis under buffered conditions the deoxyribonucleoside H-phosphonates 34 (Scheme 6) in yields ranging from 77-90% (Froehler et al., 1986).
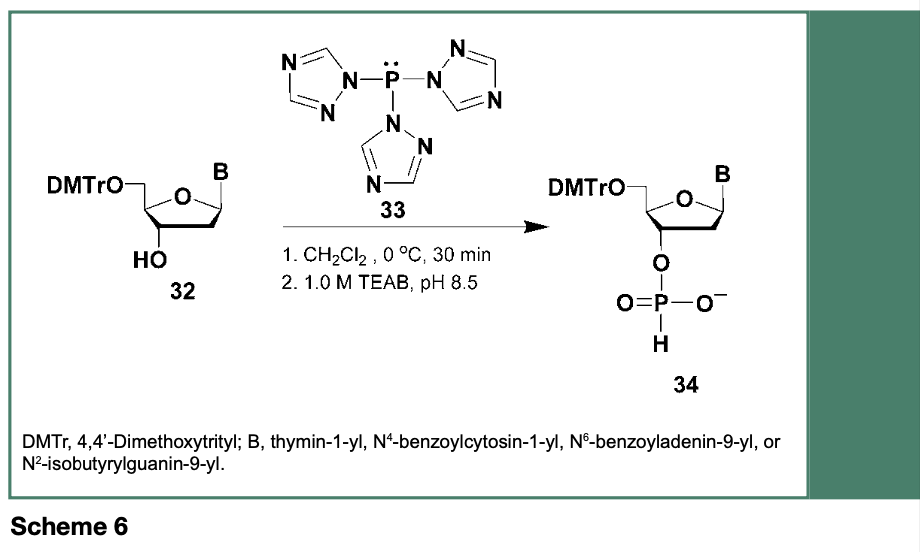
The use of 34 toward the automated synthesis of oligodeoxyribonucleotides has been demonstrated to proceed upon activation of 34 by acyl chlorides such as trimethylacetyl chloride (Froehler et al., 1986) or adamantoyl chloride (Andrus et al., 1988) to generate, as shown in Scheme 7, the reactive intermediate 35, which rapidly reacts with a 3’-O-protected ribonucleoside (36) to provide the dinucleoside H-phosphonate 37.
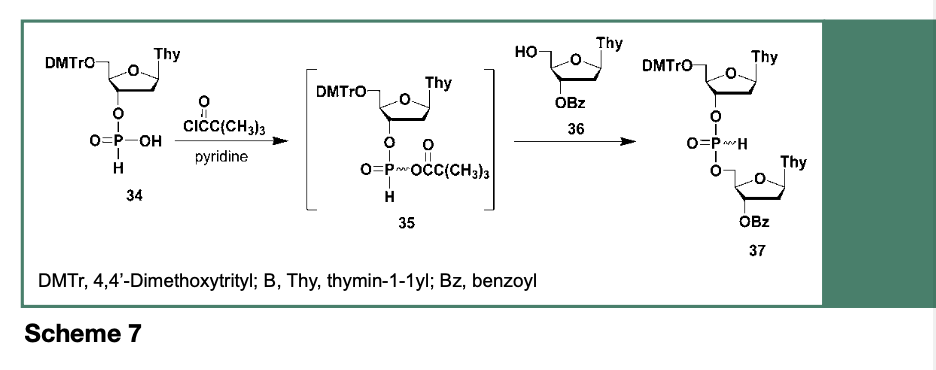
While the use of deoxyribonucleoside H-phosphonates has led to the solid-phase synthesis of a relatively large oligodeoxyribonucleotide (107-mer, Froehler et al., 1986); this synthetic strategy was not without problems. The most detrimental reaction to contend with, was the aqueous oxidation
of polynucleoside H-phosphonates that is required to produce the desired DNA sequence with native phosphodiester functions. Alkaline hydrolysis of H-phosphonate diester linkages led to significant chain cleavage, which contributed to decrease the efficiency of the H-phosphonate approach to the synthesis of DNA sequences.
Although the phosphoramidite approach continues to be preferred for solid-phase synthesis of DNA sequences based on higher stepwise yields and fewer side-products, the H-phosphonate approach is particularly well-suited for the synthesis of RNA and modified DNA sequences (Stawinski and Krazewski, 1998; Strömberg and Stawinski, 2004). More recently, the H-phosphonate chemistry has found applications in the synthesis of DNA and RNA analogs including: (i) boranealkylphosphines (Roy and Caruthers, 2013), boranephosphonates and metallophosphonates; (ii) the solid-phase synthesis of phosphate/boranophosphate chimeric DNA sequences (Sato et al., 2019); and (iii) the stereocontrolled synthesis of boranophosphate DNA sequences (Hara et al., 2019). Furthermore, the H-phosphonate chemistry has been comprehensively reviewed to include the synthesis of neutral and charged antiviral and anticancer pronucleotides (Krazewski et al., 2020).
As mentioned above, the currently established chemical strategies for solid-phase synthesis of DNA and RNA sequences take advantage of the reactivity of nucleoside phosphoramidites to form nucleic acid biopolymers, theoretically enabling the high-throughput synthesis of polynucleotides of any desired sequences. Despite the sequence-engineering versatility and the solid-phase advantages inherent to phosphoramidite-based synthesis, the multiple single-nucleotide-extension- per-synthesis and template-independent nature of the methodology poses serious challenges to product purity, particularly during pharmaceutical development at commercial scale, from which, contaminant by-products become a significant safety issue.
Minimizing the formation of process-related impurities during solid-phase synthesis of DNA and RNA sequences as potential nucleic acid-based drugs
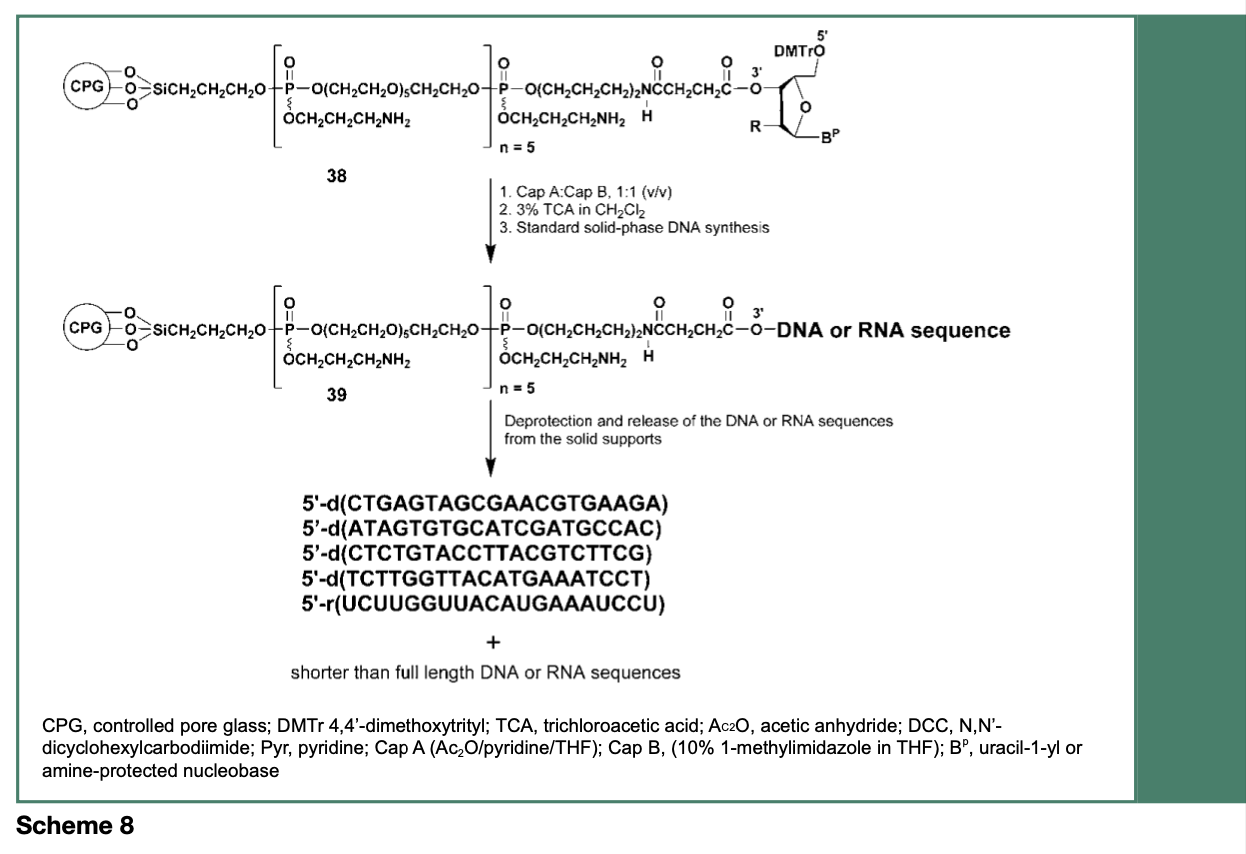
Synthetic nucleic acid products shorter than the desired full-length sequence, by as little as one nucleotide (n-1), have been reported as an appreciable contaminant in phosphoramidite-based procedures. Such shorter (n-1) heterogenous contaminant sequences can be particularly difficult to remove from a desired full-length product of clinical interest; these contaminants can potentially elicit immune responses and adverse events might arise from off-target activities upon administration to patients. Interestingly, about 45% of all shorter-than- full-length polynucleotide sequences are produced during the first four solid-phase synthesis cycles, with little truncation of product occurring during the final synthesis cycles (Temsamani, Kubert & Agrawal, 1995). Thus, it can be argued that the physical proximity between the nascent biopolymers covalently bound to the commercial controlled- pore glass (CPG) support that is usually used for polynucleotide syntheses, might interfere with the free diffusion of reagents during early reaction cycles.
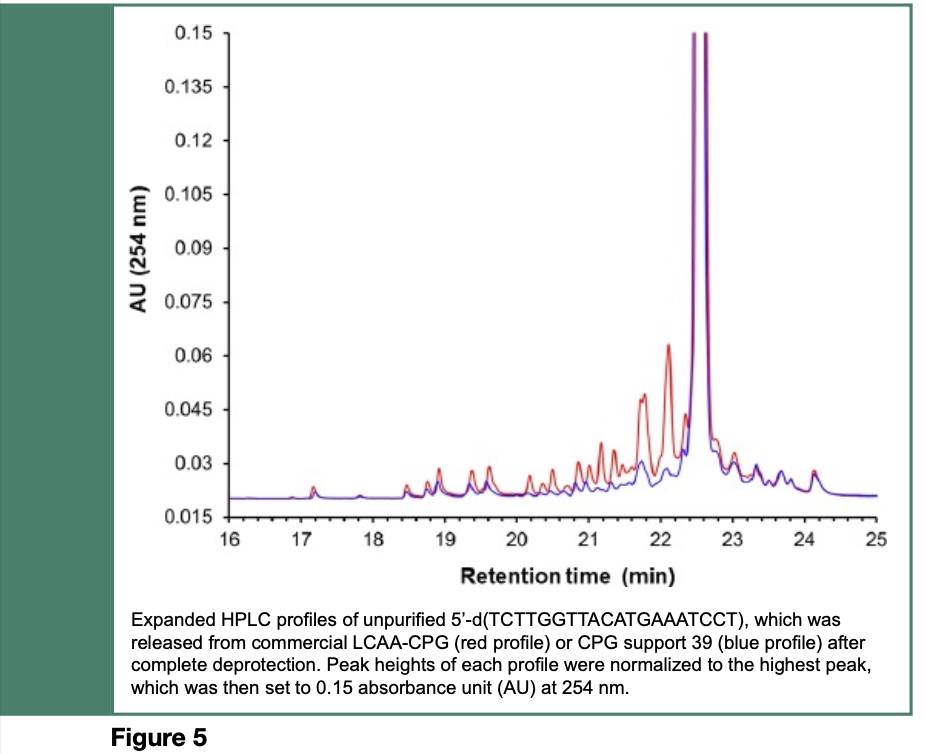
As illustrated in Scheme 8 and Figure 5, modification of the CPG support with multiple hexaethylene glycol spacers, has resulted in a reduction of the amounts of process-related impurities in synthetic nucleic acid sequences by more than 40%, when compared with the same sequences made from an unmodified commercial CPG support. Although such a reduction of process-related impurities is substantial, complete removal of shorter sequence contaminants from the full-length nucleic acid sequence has yet to be achieved.
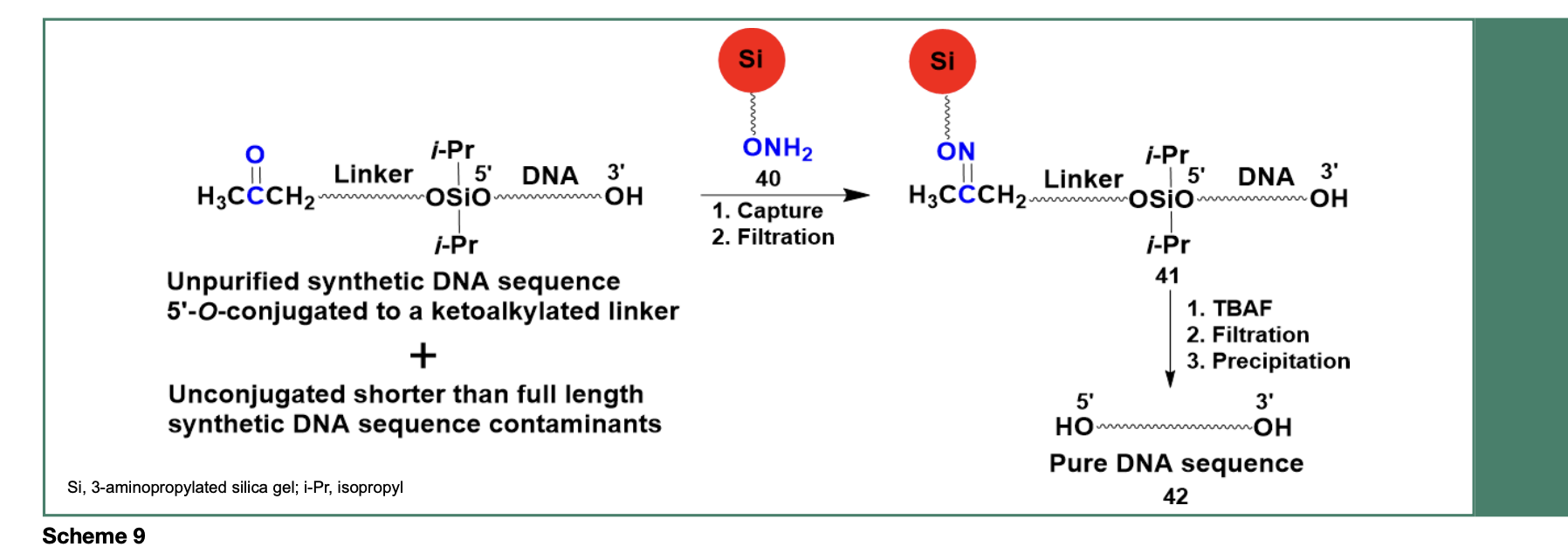
An improved method for the purification of DNA sequences had to be developed and implemented to achieve near quantitative (~98%) removal of shorter sequence contaminants. As presented in Scheme 9, a method consisting of an aminooxylated capture support (40), capable of chemoselectively capturing an unpurified DNA sequence through an efficient oximation reaction, yielded the solid-phase-captured
DNA sequence 41 while unbound shorter than full length DNA sequence contaminants were washed off, near quantitatively, from the capture support 41.
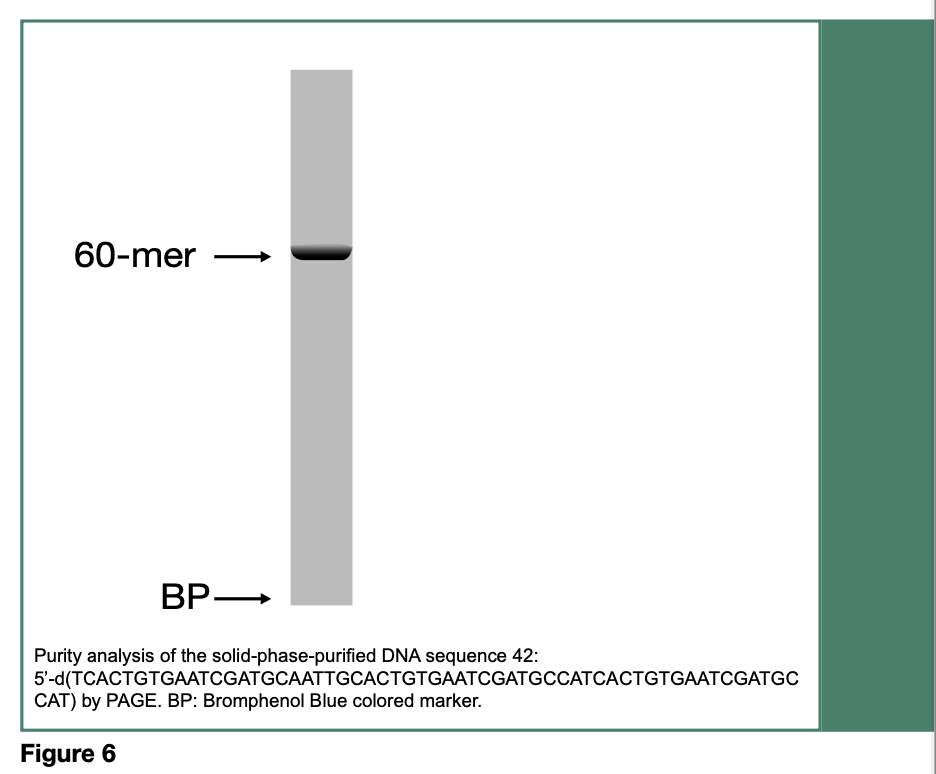
Release of the desired DNA sequence from 41 is achieved upon treatment with tetra-n-butylammonium fluoride, followed by filtration and precipitation of the DNA sequence 42 in dry tetrahydrofuran. The purity of 42 (60- mer) has been assessed by polyacrylamide gel electrophoresis (PAGE, Figure 6) and measured to be 98%, based on RP-HPLC analysis of the isolated DNA sequence.
In addition to shorter than full length DNA sequence contaminants, as process-related impurities, one must take into consideration that oligoribonucleotides have become an important class of therapeutic biomolecules, despite the daunting challenge associated with protecting an additional ribonucleoside 2’-hydroxyl function, which is absent in deoxyribonucleosides.
To prevent unwanted reactions with the 2’-hydroxyl of ribose moieties, the search for an optimal 2’-hydroxyl protecting group for ribonucleosides has been ongoing for essentially half a century; the protecting group must remain stable during RNA chain assembly and, for most synthetic strategies, be the last to be cleaved during deprotection of the synthetic RNA sequences. Cleavage of the protecting group must also be quantitative under conditions that will not induce cleavage or migration of internucleotide linkages.
Thus, the use of monomeric 2’-O-acetal- protected ribonucleoside phosphoramidites for solid-phase synthesis of RNA sequences began in the early 2000s and, eventually, ribonucleosides with a flexible 2’-O-iminooxymethyl acetal function came to be of particular interest for two reasons: (i) first, deprotection could be effectuated under neutral conditions (thereby avoiding RNA- labile extremes of pH during deprotection); (ii) second, a stable protection of the 2’-hydroxyl is maintained under the basic conditions necessary for deprotection of the nucleobase and phosphate-protecting groups. Although the development of 2’-O-acetal-protecting groups has been an important milestone; several 2’-O-acetal protecting groups are notorious for generating formaldehyde, as an untenable mutagenic by-product (Liber et al., 1989; Merk and Speit, 1998; Wilson et al. 2019) created during deprotection of synthetic RNA sequences. Moreover, 2’-O-acetal protecting groups requiring fluoride ions for acetal cleavage, produced damaging RNA alkylating side products (e.g., acrylonitrile) in addition to formaldehyde and fluorotrialkylsilane contaminants during RNA sequence deprotection. Research efforts have been made in the Beaucage lab to develop and implement a 2’-O-protecting group that can eliminate completely the production of the above side-products, when deprotecting synthetic RNA sequences.
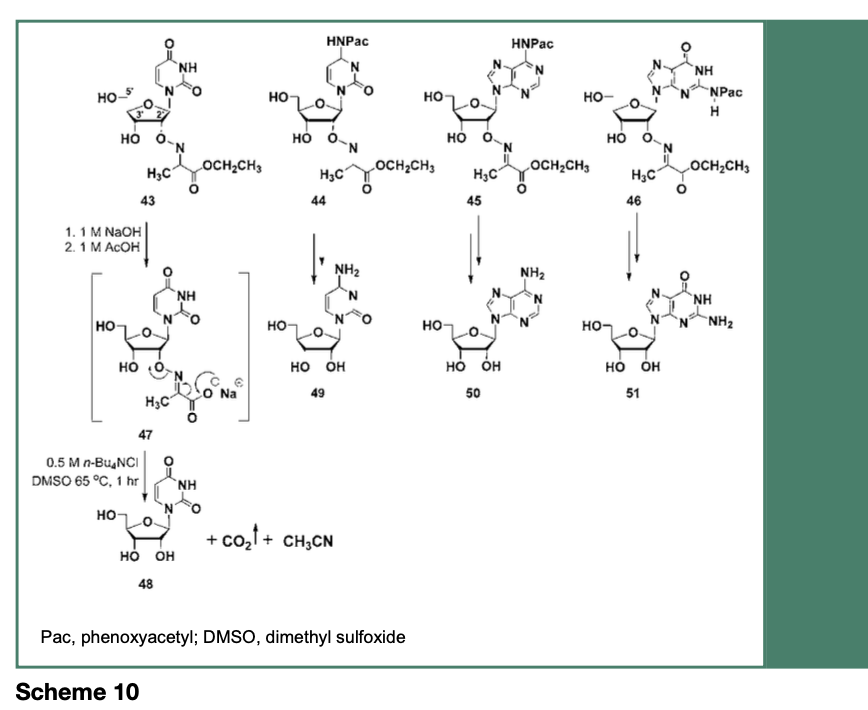
As depicted in Scheme 10, the ribonucleosides 43-46 are each protected as stable 2’-O-imino- 2-methyl propanoic acid ethyl esters through multistep chemical synthesis protocols (Takahashi et al., 2021). Although 2’-O-imino- 2-methyl propanoic acid ethyl esters are stable protecting groups for ribonucleosides, the eventual cleavage of these protecting groups has been investigated.
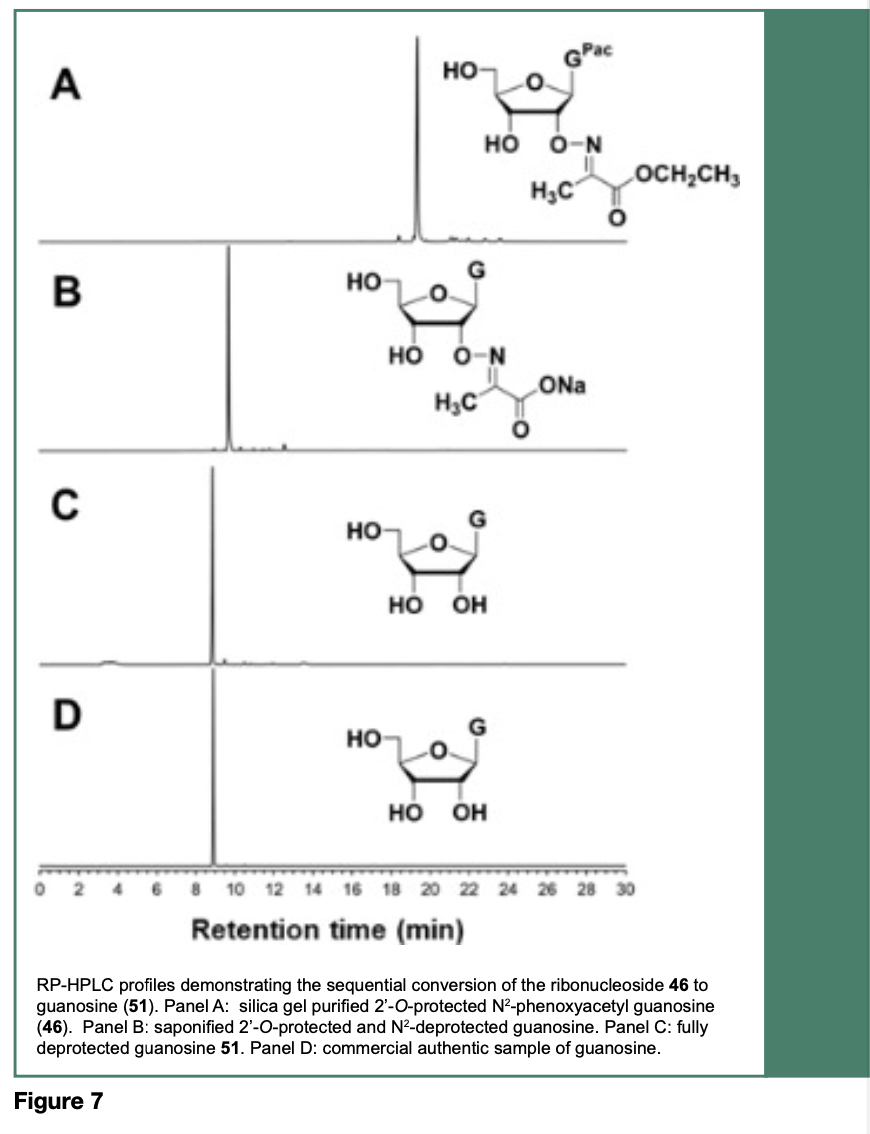
Treatment of the ribonucleoside 43-46 with aqueous 1 M sodium hydroxide resulted in the saponification of their ethyl esters and the cleavage of their N-4, N-6 or N-2-protecting groups to provide their respective 2’-O-imino- 2-propanoic acid sodium salt, which upon heating at 65 °C in a solution of 0.5 M tetra-n- butylammonium chloride (n-Bu4NCl) in DMSO, induced an intramolecular decarboxylative elimination of carbon dioxide and acetonitrile to produce the native ribonucleosides 48-51. Figure 7 specifically shows the deprotection of 46 to provide 51, which is comparable to an authentic commercial guanosine sample, based on respective reverse-phase HPLC (RP-HPLC) profiles.
Given that each ribonucleoside 2’-O-imino-2- propanoic acid ethyl ester can be efficiently deprotected, the corresponding fully protected ribonucleoside phosphoramidites 52, 53, 54 and 55 (Figure 8) were prepared and used in the solid-phase synthesis of a chimeric RNA sequence (20-mer) with the intent of comparing the coupling efficiencies of these phosphoramidites with those of commercial fully protected ribonucleoside 2’-O-TBDMS phosphoramidite monomers, when employed for solid-phase synthesis of the same RNA sequences conducted under essentially identical instrumental conditions.
Specifically, solid-phase synthesis of an RNA sequence was conducted on a succinyl long chain alkylamine controlled-pore glass (CPG) support functionalized with 2’-deoxythymidine as the leader nucleoside. The monomeric ribonucleoside phosphoramidites, 52, 53, 54 and 55 were each dissolved and used under the conditions recommended by the manufacturer of the DNA/RNA synthesizer.
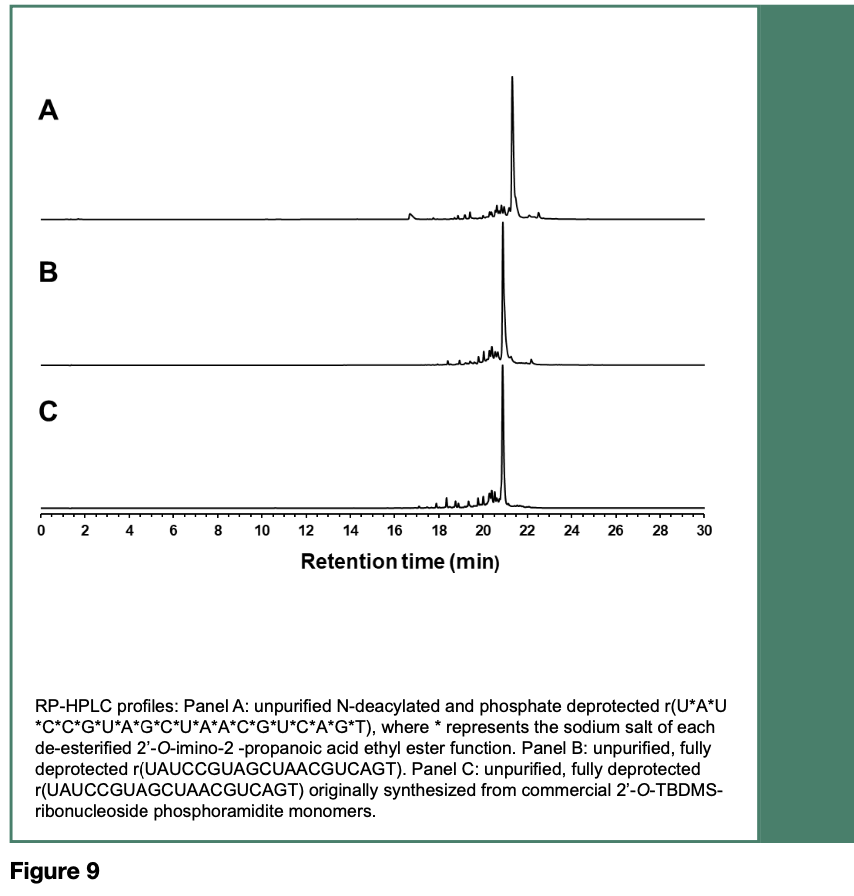
The coupling reaction time for each activated ribonucleoside phosphoramidite was set to 5 min. The RNA sequence was fully deprotected, desalted and analyzed by RP- HPLC. As depicted in Figure 9, the main peak area of the RNA sequence made from the phosphoramidites 52-55 (Panel B) accounted for 60% of the total peak areas, with phosphoramidite coupling efficiencies averaging 97% per coupling cycle; the main peak area of the same RNA sequences made from commercial 2’-O-TBDMS phosphoramidites (Panel C) accounted for 55% of the total peak areas of this pyrimidine-rich RNA sequence composed of 11 pyrimidines and 9 purines.
A purine-rich RNA sequence made of 6 pyrimidines and 14 purines had also been solid-phase synthesized and fully deprotected under rigorously similar experimental conditions to provide similar outcomes, based on the coupling efficiencies of the phosphoramidites 52-55 averaging 98% per coupling cycles.
Thus, this synthetic strategy met the objectives of: (i) minimizing the production of process- related impurities through improvements made to solid support materials for increasing the production of the full length DNA/RNA sequences while decreasing the shorter than full-length DNA/RNA sequence contaminants and (ii) eradicating the formation of alkylating/mutagenic side-products through the development and implementation of innovative 2’-hydroxyl protecting groups for RNA sequences that cannot generate such side- products, when subjected to deprotection conditions.
One can therefore conclude that the purity of synthetic nucleic acid sequences is of paramount importance to the manufacture of safe, potent and efficacious nucleic acid-based drugs.
Whether the product of interest is a DNA, or an RNA sequence intended for gene silencing strategies or a DNA sequence planned for ultimate use in gene therapy, the safe and effective application of the nucleic acid product in the clinical settings will ultimately emanate from the establishment of rigorously dependable synthetic methods. Synthetic nucleic acids, that prove in vitro to be highly potent at silencing the expression of disease-causing proteins or promoting the therapeutic expression of functional gene products, can mature as drug candidates only if they can be deemed free of process-related impurities. The work described here is the expression of a commitment to manufacture high-quality nucleic acid-based drugs to benefit humanity.


