Dr Kim Hammond-Kosack, Research Programme Leader Wheat Pathogenomics at Rothamsted Research, explores how big data analyses and knowledge networks can be used to protect global wheat crops from infectious diseases
The United Nations General Assembly has declared 2020 as the International Year of Plant Health (IYPH). The year is a once in a lifetime opportunity to raise global awareness on how protecting plant health can help end hunger, reduce poverty, protect the environment, and boost economic development. The Food and Agriculture Organization (FAO) estimates that agricultural production must rise by about 60% by 2050 to feed a larger and generally richer population.
Wheat is the number one arable crop grown in the UK and Europe. Globally wheat provides 20% of the total calories consumed by humans each day and is a valuable source of animal feed. However, the UK, EU and global wheat crops are under severe threat. For the past decade, farm yields have remained static due to adverse weather conditions, deteriorating soil health, and various other stresses to plants.
Global warming is allowing many kinds of disease-causing organisms to move beyond their typical ranges, which is introducing diseases into new (and potentially unprepared) areas. Collectively, disease-causing microbes affect all parts of the plant and can reduce potential wheat yields by up to 25%. Some wheat diseases of particular concern include stem rust, wheat blast, powdery mildew, Septoria leaf blotch, Fusarium head blight and ‘take-all’ root disease.
Since the completion of the first whole genome sequencing projects, the amount of data created from genome sequencing now doubles every seven months. By 2020, data will be generated a million times faster than it was ten years ago. The biosciences has entered the ‘post-genomic era’, where analysing data on a massive scale is the new norm.
In order to benefit many bioscience disciplines, including plant and crop health, it is imperative to ensure that the information being produced is Findable, Accessible, Interoperable, and Reusable (following the FAIR principles). Equally important is to develop novel ways to analyse data, such as using interactive networks and artificial intelligence to go from genome to genotype to phenotype.
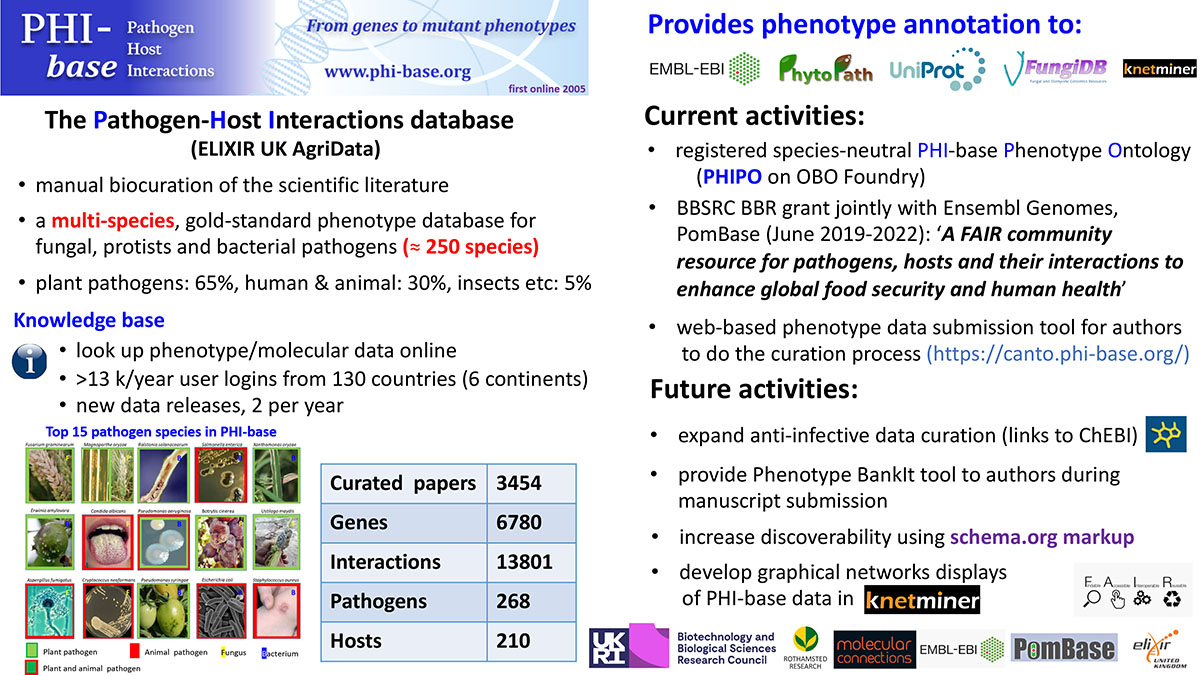
The Pathogen-Host Interactions database – PHI-base
The multispecies data resource PHI-base focuses on infectious microbes and their disease afflicted hosts (Figure). PHI-base stores gold-standard phenotypes (visible traits) on genes implicated in the disease-causing ability of a pathogen (virulence) and also the host gene products first targeted by pathogen derived mobile molecules called ‘effectors’ that enter the host and directly affect the interaction outcome. PHI-base is a primary information source for the global research community studying plant-pathogen interactions.
Manually curated information from ~3,500 research articles is accessible to researchers to easily familiarise themselves with relevant molecular and biological facts on pathogenicity, virulence and effector genes and their 1st host targets. Nine species neutral, high-level phenotypes are used to describe the overall experimental interaction outcomes enabling comparative analysis of different pathosystems. The phenotypic data in PHI-base can be directly traced to the individual gene entries within the genomes of > 300 plant pathogenic species available within specialist genome databases including ENSEMBL invertebrates and FungiDB. PHI-base adheres to the FAIR principles and is a member of the UK node of ELIXIR’s ‘Data for Life’ project, where it supplies gold-standard agricultural–genomics data.
Over the past 4 years, due to increasing concerns over global food security, crop species entries in PHI-base have increased. Currently, 37% of PHI-base interaction entries are for wheat, rice, maize, barley, tomato, potato and brassica. Wheat entries are now at 13% and cover 18 globally important pathogenic species.
Devising new ways to control plant disease
The curated phenotypes and genotypes in PHI-base can be used together with the sequenced genome information to increase the range of approaches used to control disease in wheat and other crops. These new approaches include: Tracking the genes that allow pathogens to cause disease, targeting the removal of host susceptibility genes in plant breeding programmes, undertaking pathogen effector lead direct screening of plant germplasm collections to maintain the widest possible repertoire of plant disease resistance genes in plant breeding programmes, identifying new pathogen genes that biochemical and biotechnology industries can target to suppress or kill pathogens and finally undertaking pan-genome analyses between historic and current strains of each pathogen to explore how these strains have evolved over time to overcome existing control strategies.
To try and make sense of the huge amount of biological data that is being produced, researchers are exploring new ways to visualise and combine data, with the ultimate aim of finding (and exploiting) new weaknesses in plant pathogens. One of the most powerful tools to be developed for plants and their associated pathogens is Knetminer.
Knetminer will allow researchers without specialist bioinformatics skills to explore a wealth of existing genomic information from multiple species and compare this with their own experimental data to permit rapid progress, new insights and new discoveries. Knetminer connects information from databases and the literature in an intelligent data model – known as a knowledge graph – that understands biological entities and their relationships to one another as concepts, rather than raw text.
Knetminer enables scientists to search the knowledge graph for genes, phenotypes, stresses, molecules and other information – and instantly tells the stories of complex traits and diseases for both pathogens and their hosts using explainable artificial intelligence. The combination of knowledge graphs, artificial intelligence and digital technologies will transform the way researchers work, and help to protect crops from microbial pathogens more efficiently and sustainably.
References
https://www.ippc.int/en/iyph/
https://doi 10.1111/mpp.12618
https://doi 10.1038/sdata.2016.18
https://doi: 10.1093/nar/gkx1011
https://doi: 10.1016/j.gde.2013.10.003
https://doi 10.1126/science.aar7191
https://doi 10.1093/nar/gkw1089
http://www.phi-base.org/
https://en.wikipedia.org/wiki/PHI-base
https://doi 10.1093/nar/gkv1052
https://doi 10.1093/nar/gkz890
https://bit.ly/350Hm1P
https://zenodo.org/record/3471776#.XZY8z0Yzbcs
https://doi 10.3389/fmicb.2019.02721
https://doi 10.1016/j.atg.2016.10.003
https://knetminer.org
Please note: This is a commercial profile