Jean-Pierre Leburton, and Olgica Milenkovic, Professors of Electrical and Computer Engineering at the University of Illinois at Urbana-Champaign, discuss how big data processing via bio-sensing, and blue energy production revolutionise solid-state nanopores
Ion-channels as nanopores for DNA sequencing
Porous materials were well-known for their applications in liquid filtering and water desalination when J. Kasianowicz and his collaborators proposed to use ion-channels in the lipid bi-layer membranes of human cells as a cheap tool for reliable DNA sequencing [1]. With this methodology, now implemented in the MinION portable device developed by Oxford Nanopore Technology, a biological membrane separates two electrolytic reservoirs or sub-cells experiencing an electric potential difference that generates an ionic current through the ion-channel or nanopore.
DNA molecules guided into one of the sub-cells are then threaded through the nanopore by the electric field, blocking the ion flow with an intensity related to the presence of a particular base in the pore, thereby reading the sequence of nucleotides. This approach has its drawbacks as it is limited to the natural nanopore geometry and is vulnerable to
harsh (e.g., thermal and mechanical) surrounding conditions.
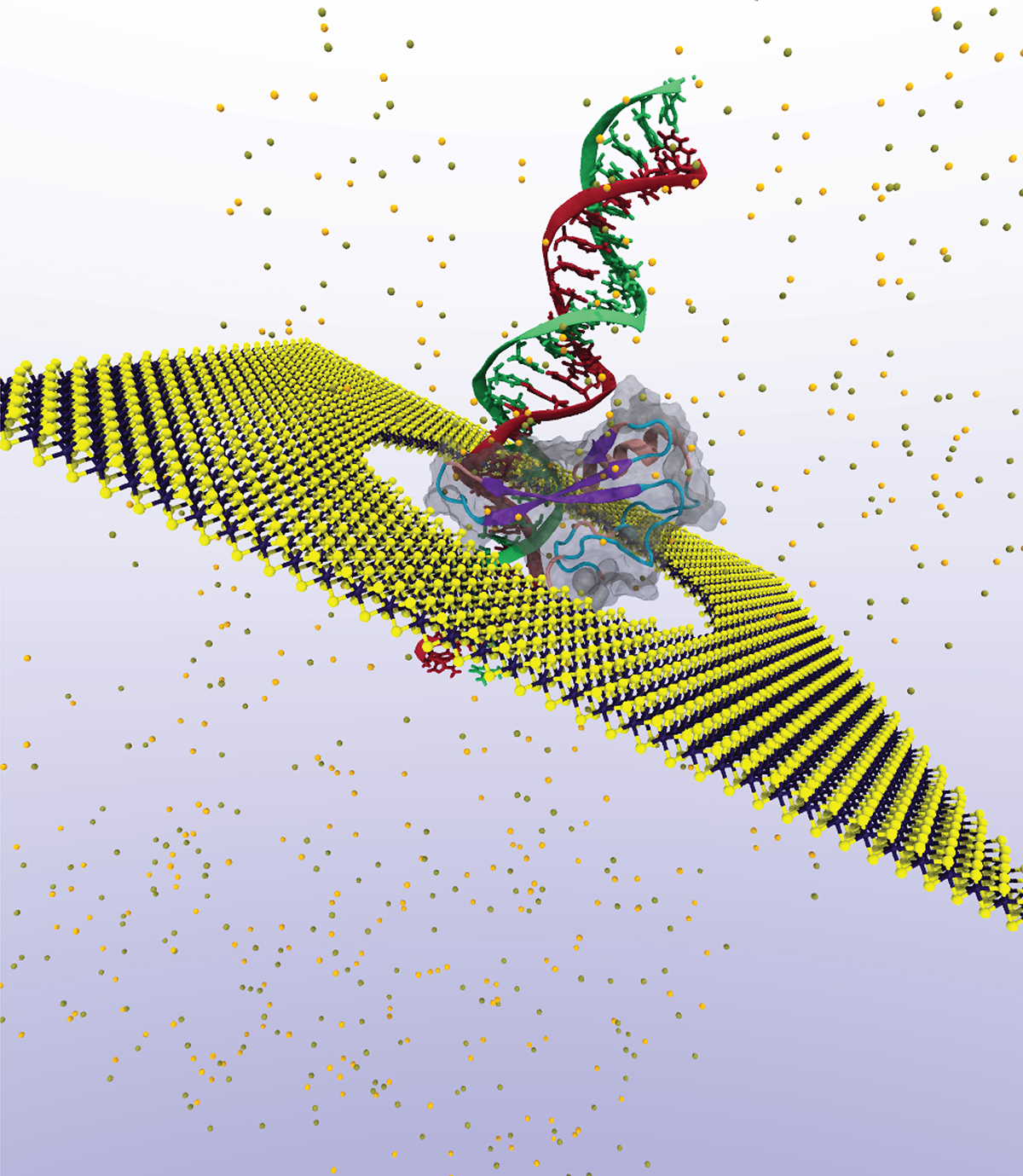
Solid state nanopores: What are they?
Coincidentally, advances in lithography techniques and semiconductor nanotechnology have enabled the fabrication of a few atom-thick solid-state membranes containing etched pores of nanometer feature sizes, which provides the physical ingredients for mimicking bio-chemical processes through cellular ion-channels with inorganic materials. Because of their synthetic nature, solid-state nanopores offer technological advantages compared to biological ion channels, not only in terms of robustness to the environment, but also in their ability to be tailored towards specific applications by a-priori designing their geometry and diameter.
Asides from DNA sequencing, these pores can be used for high-resolution sequencing of larger molecules, such as proteins attached to methyl groups along DNA strands that are epigenetic modifications heavily involved in gene expression and regulation critical to the progression of diseases such as cancer [2].
Nanopores versatility in 2D materials:
Recently discovered two-dimensional (2D) materials such as graphene or transition metal dichalcogenides (TMD) with an atomic-size thickness comparable to the DNA base separation open new avenues in nucleotide discrimination and identification with nanopore technologies. They also augment their functionality, as demonstrated by M. Graf et al., who by shining light on MoS2 membranes generate surface charges that boost their osmotic power to record high-power outputs in alkaline solutions [3]. This new discovery has promising applications in desalination, and in blue energy conversion if placed in river estuaries, for instance.
Multi-layer membrane engineering:
One of the major advantages of the 2D solid-state technology resides in the electrical, mainly semiconducting properties of the material themselves, which add another degree of freedom to the nanopore detection process by running an electronic current along the membrane. Hence, narrowing the membrane around the nanopore creates a resistance involving the biomolecule that translocates through the pore. Its detailed structure can then be readily detected by reading the membrane conductance variations.
Moreover, the ability to stack 2D materials layers by van der Waals epitaxy greatly enhances nanopore sensitivity by separately functionalizing each individual layer for a specific task as in a field-effect transistor membrane [4].
Solid state nanopores for Big Data storage and processing:
These kinds of membrane devices have recently received significant attention for use in the emerging area of DNA-based and molecular data storage and computing. In [5], the first experiments demonstrating the actual capability of ON devices to read user-defined image content encoded in synthetic DNA were reported, while other works have since demonstrated that native and solid-state nanopores can also be successfully employed for reading information encoded in polymers, proteins, chemically modified DNA bases designed for the purpose of increasing the coding alphabet, and topological modifications, such as nicks or toeholds in the sugar-phosphate backbone of DNA. The latter storage architectures required new design principles for nanopores and machine learning and signal processing techniques for raw current readouts.
Even in the presence of a significant amount of base-calling errors, often in excess of 10% of the stored content, specially designed encoding strategies involving balancing and so-called trace reconstruction processes, along with tailor-made alignment algorithms, enabled error-free reconstruction of the data.
The aforementioned works have also shown that one can jointly optimise the design of the nanopore readout channel and the data alphabet and has lead to the creation of a new area in coding theory concerned with specialised indel models and coded trace reconstruction mechanisms.
References
[1] J.J. Kasianowicz, E. Brandin, D. Branton, and D.W. Deamer. “Characterization of individual polynucleotide molecules using a membrane channel”. PNAS USA. 93 (1996):13770-13773.
[2] H. Qiu, A. Sarathy, K. Schulten and J.P.Leburton “Detection and mapping of DNA methylation with 2D material nanopores”. npj 2D Materials and Applications, 1, (2017):3
[3] M. Graf, M. Lihter, D. Unuchek, A. Sarathy, J.P. Leburton, A. Kis and A. Radenovic. “Light-Enhanced Blue Energy Generation Using MoS2 Nanopores.” Joule 3 (2019) : 1549-1564.
[4] A. Girdhar, C. Sathe, K. Schulten and J. P. Leburton. “Graphene quantum point contact transistor for DNA sensing.” PNAS USA. 110 (2013) : 16748-16753.
[5] S.M. Hossein Tabatabaei Yazdi, R. Gabrys, and O. Milenkovic. “Portable and error-free DNA-based data storage.” Scientific reports 7, no. 1 (2017): 1-6.
Please note: This is a commercial profile
© 2019. This work is licensed under CC-BY-NC-ND.











