Open Access Government produces compelling and informative news, publications, eBooks, and academic research articles for the public and private sector looking at health, diseases & conditions, workplace, research & innovation, digital transformation, government policy, environment, agriculture, energy, transport and more.
Home 2025
Archives
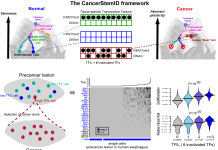
Predicting cancer risk with computational biology
Andrew Teschendorff, Professor at the Chinese Academy of Sciences, is developing computational systems-biological tools to identify cells at risk of turning cancerous.