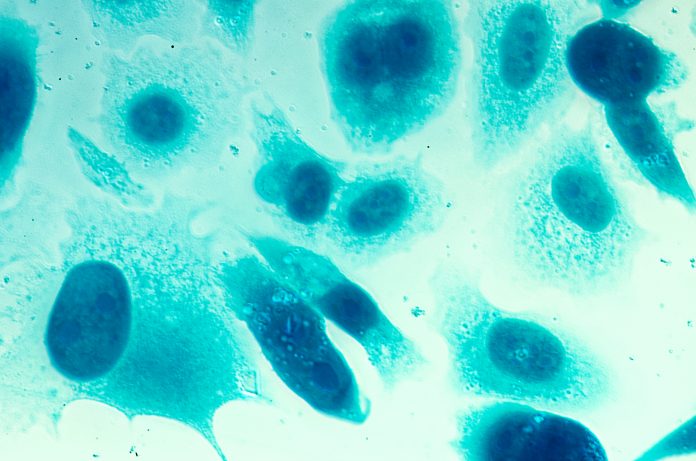
Sabine Mai and Aline Rangel-Pozzo from CancerCare Manitoba Research Institute, The University of Manitoba, explore how 3D imaging and quantitative analysis provide key insights into genomic instability
Genomic instability is a hallmark of cancer. It drives the evolution of cancer cells having a genome that differs from that of normal cells. It enables the development of cell clones and cell-to-cell heterogeneity within the tumour and between the tumour and its metastatic site(s).
The presence of genomic instability in cancer was first reported by David von Hanseman (1858-1920). His work as a pathologist in Virchow’s group in Berlin concluded that cells exhibiting genomic instability were not found in normal tissue but only in tumour tissues. His illustrations showed chromosomal instability, which included aberrant chromosome numbers (aneuploidy: gain or loss of chromosomes); and chromosomal missegregation between daughter cells.
The first mechanistic studies into genomic instability were performed by Theodor Boveri (1862-1915), whose work proposed that genomic instability led to the development of cancer and was not compatible with normal cell life. While he did not use the term ‘genomic instability’, his data clearly implicated it in this malignant process.
Genomic instability is a dynamic process and continues to evolve with every cell division. Therefore, if samples are taken at an early time point of tumour development and at later stages, genetic changes will have occurred.
The three-dimensional (3D) space of the nucleus: spatial genome disorder in cancer
The nucleus harbours genetic information. This information is not simply organised in a random manner but has a clear organisation. In normal cells, chromosomes assume specific places within the 3D space of the nucleus. The spaces occupied by the chromosomes are called ‘chromosome territories’. They have been conserved during evolution, and they are cell-type and differentiation-specific. This organisation has functional importance; for example, animals with night vision have a chromosome organisation in their rod cells that is distinct from the one found in animals with diurnal vision. Similarly, hepatocytes have a different arrangement of chromosome territories and chromosome neighbourhoods than lymphocytes.
The organisation of chromosomes and of the genes they harbour is key to transcriptional regulation. Genes located near the periphery of the 3D nucleus are commonly repressed, while genes found in the centre of the nucleus are expressed. Cancer cells have re-ordered the genetic information within the nuclear space and often change the position and orientation of chromosomes, which enables new transcription patterns. Cancer cells also change the way their DNA is packaged and increase the presence of interchromatin space. Moreover, when using telomeres, the ends of chromosomes, as structural markers for genome (in)stability, cancer cells, compared to normal cells, show very significant changes in their 3D organisation.
3D telomere profiling in cancer
3D imaging of telomeres developed and carried out at the Genomic Centre for Cancer Research and Diagnosis (GCCRD) at the University of Manitoba and CancerCare Manitoba (Winnipeg, Canada) enabled the fine mapping of nuclear organisation of the cancer cell genome. At nanoscale resolution, Dr Mai’s team and collaborators visualised and measured the structural organisation of the cancer genome; telomeres were used as surrogate markers of genome organisation and proved to play a role as structural biomarkers of genomic instability.
Multiple cancers were examined. 3D telomere profiling allowed the team to quantify the level of genomic instability, the risk to progression and/or the response to treatment. Examples include lymphoid (such as Hodgkin’s Lymphoma, Multiple myeloma, Chronic Myeloid Leukemia, Myelodysplastic Syndromes and Acute Myeloid Leukemia) and solid tumours (such as neuroblastoma, glioblastoma, thyroid cancer, and prostate cancer). The software developed by the team (TeloView® – currently proprietary to Telo Genomics Corp. Toronto, Canada) allowed for single-cell profiling and informed about each patient’s cancer.
As a result of these initial clinical studies, a biotech company, Telo Genomics Corp., was founded. Telo Genomics Corp. is located at the MaRS Discovery District in Toronto, ON, Canada (telodx.com). The company is dedicated to personalised medicine solutions in cancer, with a current focus on multiple myeloma.
Lamin A/C in cancer
Lamin A/C is a protein found in the nucleus of the cells. It has many roles which include maintenance of the correct cell shape and regulation of gene activity, i.e., when genes will be switched on during the cell’s lifetime. To maintain structural cell stability, lamin A/C increases nuclear firmness while still allowing flexibility, which helps to maintain proper nuclear structure even under mechanical tension. Nuclei with mutations in lamin A/C genes, tend to be more fragile and to misshape easily under mechanical stress. Moreover, complete loss of lamin A/C has been shown to be lethal in mice. It is also known that cancer cells can deregulate lamin A/C expression, which can result in less or more proteins produced by the cells. However, due to the high overall variability in expression levels within cancer types and lamin A/C correlation with various important signalling pathways in the cell, lamin A/C’s role in cancer development and progression still waits for future clarification.
The Mai group investigated, for the first time, at the single-cell level, the 3D expression pattern of lamin A/C protein in Classical Hodgkin’s lymphoma (cHL) patients’ samples and in patient-derived cell lines, in comparison to peripheral blood lymphocytes (PBLs). cHL cell lines presented with higher levels of lamin A/C than normal PBLs. Interestingly, high levels of lamin A/C in cHL cells led to the formation of internal lamin A/C structures. Furthermore, in multiple myeloma cell lines, the downregulation of lamin A/C moved some chromosomes back to their original territories, similar to the nuclear spatial organisation found in normal B lymphocytes. These findings highlight the importance of lamin A/C expression investigation and spatial distribution through single-cell analysis. Such an approach can reveal disease heterogeneity and elucidate the mechanisms behind lamin A/C expression variability in cancer.
Cancer cell analysis, using structured illumination microscopy
Super-resolution microscopy techniques are critical tools in cancer research, used to investigate, in detail complex structures in the cell not evident by conventional wide-field
fluorescence microscopy. Clinically relevant and quantifiable imaging parameters are urgently needed for personalised medicine to select patients with poor prognoses for more appropriate therapy. 3D Structured Illumination Microscopy (3D SIM) enables the visualisation of structures at the nanoscale, which is ideal to investigate chromatin organisation.
The Mai team was the first to apply this technology to the study of the cancer cell genome. The goal was to compare the nuclear DNA organisation of normal cells to premalignant and malignant cells of different cancer types. The team identified that nuclei of cancer cells have more regions of empty spaces (spaces voided of DNA structure) compared to normal cells, which was named “DNA-poor spaces”, also called interchromatin spaces. The team further developed tools and quantified the organisation of nuclear DNA (using a measurement called granulometry) and applied it to the acquired super-resolution images. This technique was able to provide insights into cancer cell heterogeneity and DNA structural alterations that could lead to disease progression and treatment resistance.
Summary
The spatial organisation of the genome is key to cancer cell aggressiveness. It can be mapped in many ways using 3D imaging and quantitative analysis. Chromosome territories, lamin A/C, nuclear DNA packaging and the degree of interchromatin spaces, and 3D telomere profiling all enable insights into the dynamic structural alterations that are enabling hallmarks for cancer cell genomes.
Please note: This is a commercial profile
© 2019. This work is licensed under CC-BY-NC-ND.